Abstract
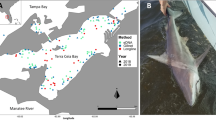
The detection of threatened species, specially where they have been depleted, is key to their conservation but poses severe logistical challenges precisely because of their very low abundances. Sawfishes (family Pristidae) are among the most threatened marine taxa with globally-reported severe reductions in population size and disappearance from large parts of their former distribution range. This study presents the first report on the use of environmental DNA (eDNA) to evaluate the presence of Critically Endangered sawfishes in parts of their former range, where no recent information about their presence exists. During 2016 and 2017 gillnet and eDNA surveys were carried out at Tecolutla Estuary, Veracruz, and Laguna de Términos and Los Petenes Biological Reserve, Campeche, in Mexico, where Pristis pectinata and Pristis pristis were abundant 40–50 years ago. Our study aimed at determining: (a) if eDNA was a suitable tool for assessing the presence of sawfishes at suspected very low abundances, and (b) if sawfishes still occur in Mexico. We found evidence of sawfish presence through eDNA samples in all three sites, at very low frequencies, but no evidence through gillnetting. Species resolution was difficulted by DNA degradation, but 2/12 samples were identified as Pristis pectinata. Our findings confirm that eDNA is a suitable tool to rapidly assess the presence of critically endangered sawfishes in the wild, stress the urgent need for eDNA surveys throughout the former range of sawfishes globally and highlight the need for swift measures to protect the remaining individuals still living in Mexico.


Similar content being viewed by others
Availability of data and materials
Data used in this study may be available upon request to the lead author.
Code availability
Not applicable.
Change history
23 April 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00227-021-03885-0
References
Bakker J, Wangensteen OS, Chapman DD, Boussarie G, Buddo D, Guttridge TL et al (2017) Environmental DNA reveals tropical shark diversity in contrasting levels of anthropogenic impact. Sci Rep 7:1–11
Barnes MA, Turner CR, Jerde CL, Renshaw MA, Chadderton WL, Lodge DM (2014) Environmental conditions influence eDNA persistence in aquatic systems. Env sci technol 48:1819–1827
Biggs J, Ewald N, Valentini A, Gaboriaud C, Dejean T, Griffiths RA et al (2015) Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol Cons 183:19–28
Bista I, Carvalho GR, Tang M, Walsh K, Zhou X, Hajibabaei M et al (2018) Performance of amplicon and shotgun sequencing for accurate biomass estimation in invertebrate community samples. Mol Ecol Resour 18:1020–1034
Bonfil R, Mendoza-Vargas OU, Ricaño-Soriano M, Palacios-Barreto PY, Bolaño-Martínez N (2017) Former widespread abundance and recent downfall of sawfishes in Mexico as evidenced by historical photographic and trophy records. Fisheries 42:256–259
Bonfil R, Ricaño-Soriano M, Mendoza-Vargas OU, Méndez-Loeza I, Pérez-Jiménez JC, Bolaño-Martínez N, Palacios-Barreto P (2018) Tapping into local ecological knowledge to assess the former importance and current status of sawfishes in Mexico. Endang Species Res 36:213–228
Boussarie G, Bakker J, Wangensteen OS, Mariani S, Bonnin L, Juhel JB et al (2018) Environmental DNA illuminates the dark diversity of sharks. Science Advan 4:eaap9661
Carlson JK, Gulak SJB, Simpfendorfer CA, Grubbs RD et al (2014) Movement patterns and habitat use of smalltooth sawfish, Pristis pectinata, determined using pop-up satellite archival tags. Aquat Conserv 24:104–117
Carranza J (1959) Pesca y Recursos Pesqueros In: E Beltrán (ed) Los Recursos Naturales del Sureste y su Aprovechamiento. III Parte, Estudios Particulares Tomo 3°. Instituto Mexicano de Recursos Naturales Renovables, A C, Mexico DF, p 151–238
Collins RA, Wangensteen OS, O’Gorman EJ, Mariani S, Sims DW, Genner MJ (2018) Persistence of environmental DNA in marine systems. Commun Biol 1:185
Cristescu ME, Hebert PD (2018) Uses and misuses of environmental DNA in biodiversity science and conservation. Annu Rev Ecol Evol Syst 49:209–230
Davison PI, Creach V, Liang WJ, Andreou D, Britton JR, Copp GH (2016) Laboratory and field validation of a simple method for detecting four species of non-native freshwater fish using eDNA. J Fish Biol 89:1782–1793
Deiner K, Walser JC, Mächler E, Altermatt F (2015) Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biol Cons 183:53–63
Deiner K, Fronhofer EA, Mächler E, Walser JC, Altermatt F (2016) Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat commun 7:1–9
Deiner K, Bik HM, Machler E, Seymour M, Lacoursiere-Roussel A, Altermatt F et al (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol Ecol 26:5872–5895
Dejean T, Valentini A, Miquel C, Taberlet P, Bellemain E, Miaud C (2012) Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. J Appl Ecol 49:953–959
Dejean T, Valentini A, Duparc A, Pellier-Cuit S, Pompanon F, Taberlet P, Miaud C (2011) Persistence of environmental DNA in freshwater ecosystems. PLoS ONE 6:e23398
Díaz-Jaimes P, Bonfil R, Palacios-Barreto P, Bolaño-Martinez N, Bayona-Vásquez NJ (2018) Mitochondrial genome of the critically endangered smalltooth sawfish Pristis pectinata from Veracruz. Mexico Conserv Genet Res 10(4):663–666
Dulvy NK, Davidson LNK, Kyne PM, Simpfendorfer CA et al (2016) Ghosts of the coast: global extinction risk and conservation of sawfishes. Aquat Conserv 26:134–153
Elbrecht V, Leese F (2015) Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass-sequence relationships with an innovative metabarcoding protocol. PLoS ONE 10:e0130324
Evans NT, Shirey PD, Wieringa JG, Mahon AR, Lamberti GA (2017) Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 42:90–99
Faria VV, Mcdavitt MT, Charvet P, Wiley TR et al (2013) Species delineation and global population structure of Critically Endangered sawfishes (Pristidae). Zool J Linn Soc 167:136–164
Ficetola GF, Taberlet P, Coissac E (2016) How to limit false positives in environmental DNA and metabarcoding? Mol Ecol Resour 16:604–607
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4:423–425
Foote AD, Thomsen PF, Sveegaard S, Wahlberg M, Kielgast J, Kyhn LA et al (2012) Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE 7:e0041781
Furlan EM, Gleeson D (2016) Improving reliability in environmental DNA detection surveys through enhanced quality control. Mar Freshw Res 68:388–395
Goldberg CS, Strickler KM, Fremier AK (2018) Degradation and dispersion limit environmental DNA detection of rare amphibians in wetlands: Increasing efficacy of sampling designs. Sci Total Environ 633:695–703
Goldberg CS, Strickler KM, Pilliod DS (2015) Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biol Conserv 183:1–3
Goldberg CS, Sepulveda A, Ray A, Baumgardt J, Waits LP (2013) Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshw Sci 32:792–800
Harrison LR, Dulvy NL (eds) (2014) Sawfish: a global strategy for conservation. IUCN Species Survival Commission’s Shark Specialist Group, Vancouver
Hebert PD, Cywinska A, Ball SL, Dewaard JR (2003) Biological identifications through DNA barcodes. P Roy Soc Lond B Bio 270:313–321
Hering D, Borja A, Jones JI, Pont D, Boets P, Bouchez A et al (2018) Implementation options for DNAbased identification into ecological status assessment under the European water framework directive. Water Res 138:192–205
Hinlo R, Gleeson D, Lintermans M, Furlan E (2017) Methods to maximise recovery of environmental DNA from water samples. PLoS ONE 12:e0179251
Hollensead LD, Grubbs RD, Carlson J, Bethea DM (2016) Analysis of fine-scale daily movement patterns of juvenile Pristis pectinata within a nursery habitat. Aquatic Conserv Mar Freshw Ecosyst 26:492–505
Huerlimann R, Cooper MK, Edmunds RC, Villacorta-Rath C, Le Port A, Robson HL, Strugnell JM, Burrows D, Jerry DR. (2020) Enhancing tropical conservation and ecology research with aquatic environmental DNA methods: an introduction for non-environmental DNA specialists. Anim Conserv 23:632–645
Hunter ME, Meigs-Friend G, Ferrante JA, Kamla AT, Dorazio RM, Diagne LK et al (2018) Surveys of environmental DNA (eDNA): a new approach to estimate occurrence in Vulnerable manatee populations. Endang Species Res 35:101–111
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv Lett 4:150–157
Lafferty KD, Benesh KC, Mahon AR, Jerde CL, Lowe CG (2018) Detecting southern California’s white sharks with environmental DNA. Front Mar Sci 5:355
Last P, Naylor G, Séret B, White W, de Carvalho M, Stehmann M (eds) (2016) Rays of the World. CSIRO publishing and Cornell University Press, Ithaca
Lacoursière-Roussel A, Rosabal M, Bernatchez L (2016) Estimating fish abundance and biomass from eDNA concentrations: variability among capture methods and environmental conditions. Mol Ecol Resour 16:1401–1414
Lehman, RN, Poulakis GR, Scharer RM, Schweiss KE, Hendon JM, Phillips NM (2020) An environmental DNA tool for monitoring the status of the Critically Endangered Smalltooth Sawfish, Pristis pectinata, in the Western Atlantic. Conserv Genet Resour, 1–9.
Le Port A, Bakker J, Cooper MK, Huerlimann R, Mariani S (2018) Environmental DNA (eDNA): a valuable tool for ecological inference and management of sharks and their relatives. In: Carrier JC, Heithaus MR, Simpfendorfer CA (eds) Shark Research: emerging technologies and applications for the field and laboratory. CRC Press, Boca Raton, pp 255–284
Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V et al (2013) A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool 10:34
MacCall AD (1990) Dynamic geography of marine fish populations. Washington Sea Grant Program, Seattle
Moushomi R, Wilgar G, Carvalho G, Creer S, Seymour M (2019) Environmental DNA size sorting and degradation experiment indicates the state of Daphnia magna mitochondrial and nuclear eDNA is subcellular. Sci Rep 9:e12500
Mulligan CJ (2005) Isolations and analysis of DNA from archaeological, clinical, and natural history specimens. Method Enzymol 395:87–103. https://doi.org/10.1016/S0076-6879(05)95007-6
Murakami H, Yoon S, Kasai A, Minamoto T, Yamamoto S, Sakata MK et al (2019) Dispersion and degradation of environmental DNA from caged fish in a marine environment. Fish Sci 85:327–337
Naylor GJP, Caira JN, Jensen K, Rosana KAM, White WT, Last PR (2012) A DNA sequence-based approach to the identification of shark and ray species and its implications for global elasmobranch diversity and parasitology. AMNH Bull 367:263p
Piaggio AJ, Engeman RM, Hopken MW, Humphrey JS, Keacher KL, Bruce WE, Avery ML (2014) Detecting an elusive invasive species: a diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Mol Ecol Resour 14:374–380
Piggott MP (2016) Evaluating the effects of laboratory protocols on eDNA detection probability for an endangered freshwater fish. Ecol Evol 6:2739–2750
Port JA, O’Donnell JL, Romero-Maraccini OC, Leary PR et al (2016) Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Mol Ecol 25:527–541
Poulakis GR, Grubbs RD (2019) Biology and ecology of sawfishes: global status of research and future outlook. Endang Species Res 39:77–90
Poulakis GR, Stevens PW, Timmers AA, Stafford CJ, Simpfendorfer CA (2013) Movements of juvenile endangered smalltooth sawfish, Pristis pectinata, in an estuarine river system: use of non-main-stem river habitats and lagged responses to freshwater inflow-related changes. Environ Biol Fish 96:763–778
Rees HC, Maddison BC, Middleditch DJ, Patmore JR, Gough KC (2014) The detection of aquatic animal species using environmental DNA–a review of eDNA as a survey tool in ecology. J Appl Ecol 51:1450–1459
Rubinoff D (2006) Utility of mitochondrial DNA barcodes in species conservation. Conserv Biol 20:1026–1033
Sigsgaard EE, Carl H, Møller PR, Thomsen PF (2015) Monitoring the near-extinct European weather loach in Denmark based on environmental DNA from water samples. Biol Conserv 183:46–52
Sigsgaard EE, Nielsen IB, Bach S, Lorenzen ED, Robinson DP, Knudsen SW et al (2016) Population characteristics of a large whale shark aggregation inferred from seawater environmental DNA. Nat Ecol Evol 1:e0004
Simpfendorfer CA, Kyne PM, Noble TH, Goldsbury J et al (2016) Environmental DNA detects Critically Endangered largetooth sawfish in the wild. Endang Species Res 30:109–116
Takahara T, Minamoto T, Yamanaka H, Doi H, Kawabata ZI (2012) Estimation of fish biomass using environmental DNA. PLoS ONE 7:e35868
Tavares W, da Silva Rodrigues-Filho LF, Sodré D, Souza RF, Schneider H, Sampaio I, Vallinoto M (2013) Multiple substitutions and reduced genetic variability in sharks. Biochem Syst Ecol 49:21–29
Thomsen PF, Willerslev E (2015) Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv 183:4–18
Thomsen PF, Kielgast J, Iversen LL, Møller PR, Rasmussen M, Willerslev E (2012a) Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 7:e41732
Thomsen PF, Kielgast JOS, Iversen LL, Wiuf C, Rasmussen M, Gilbert MTP et al (2012b) Monitoring endangered freshwater biodiversity using environmental DNA. Mol Ecol 21:2565–2573
Turner CR, Miller DJ, Coyne KJ, Corush J (2014) Improved methods for capture, extraction, and quantitative assay of environmental DNA from Asian bigheaded carp (Hypophthalmichthys spp). PLoS ONE 9:e114329
Weltz K, Lyle JM, Ovenden J, Morgan JA, Moreno DA, Semmens JM (2017) Applications of environmental DNA to detect an endangered marine skate species in the wild. PLoS ONE 12:e0178124
Zarur-Menez A (1962) Algunas consideraciones geobiológicas de la Laguna de Términos. Camp Rev Soc Mex Hist Nat 23:51–70
Acknowledgements
Laura Zetina-Delgadillo, Triana Arguedas-Álvarez, and Michelle Carrillo-Castañeda helped during surveys. Violeta Monserrath Andrade-González kindly prepared all maps. Special thanks to Dr. Valeria Souza Saldivar, Instituto de Ecología, Universidad Nacional Autónoma de México, who kindly lent filtering equipment and materials for field expeditions. We acknowledge the dedicated help and support of fishers during survey campaigns, including Baudelio Cruz Coronel, Wilbert Segovia, and others. SEMARNAT and CONAPESCA granted the corresponding working permits (SGPA/DGVS/07618/15; SGPA/DGVS/06740/16; PPF/DGOPA-126/16), and managers of the LPBR and the Área de Protección de Flora y Fauna Laguna de Términos facilitated entrance to and work within those natural protected areas. Survey eDNA samples were processed by Alexis Janosik at the University of Western Florida under sub-contract DUNS 053000709. The TE survey was carried out under SEMARNAT’s Programa para la Conservación de Especies en Riesgo grant PROCER/CCER/DEPC/03/2016. Support for the large majority of our study was kindly provided by a Save Our Seas Foundation Keystone Grant to RB, and additional support was received from the Marine Conservation Action Fund of the New England Aquarium. We thank the reviewers who helped improve our manuscript.
Funding
Support for the large majority of our study was kindly provided by a Save Our Seas Foundation Keystone Grant to RB; additional support was received from the Marine Conservation Action Fund of the New England Aquarium and one of the surveys was carried out under SEMARNAT’s Programa para la Conservación de Especies en Riesgo grant PROCER/CCER/DEPC/03/2016.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Research was carried following all national ethical guidelines for working on National Protected Areas as well as with endangered species, and under the cover of working permits SGPA/DGVS/07618/15 and SGPA/DGVS/06740/16; PPF/DGOPA-126/16) from Secretaría de Ambiente y Recursos Naturales (SEMARNT) and Comisión de Pesca (CONAPESCA), respectively.
Consent to participate
All authors gave their consent for this publication.
Consent for publication
All authors gave their approval to be included in this publication.
Conflicts of interest/Competing interests
The authors declare no conflict of interest or competing interests.
Additional information
Responsible Editor: T. Reusch.
Reviewers: undisclosed experts.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
In the original online publication of the article, the last author affiliation was incorrectly published. The original article has been corrected.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bonfil, R., Palacios-Barreto, P., Vargas, O.U.M. et al. Detection of critically endangered marine species with dwindling populations in the wild using eDNA gives hope for sawfishes. Mar Biol 168, 60 (2021). https://doi.org/10.1007/s00227-021-03862-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-021-03862-7