Abstract
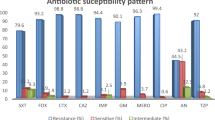
Colistin resistance in Gram negative bacteria is mainly attributed to chromosomal mutations in Two Component Systems(TCS) PhoPQ and PmrAB and plasmid-borne genes(mcr and its variants). The aim of this study was to understand the molecular basis of colistin resistance in Klebsiella pneumoniae and determine clonal transmission, in a North Indian tertiary care hospital over a 2.5 year period. Antimicrobial susceptibility was determined by Vitek and colistin resistance was confirmed by broth microdilution. Carbapenemases(blaKPC, blaVIM, blaIMP, blaNDM, blaOXA-48) and mcr-1 screening was done by PCR. Mutations in chromosomal genes mgrB, phoP, phoQ, pmrA, pmrB were analysed. Sequence typing was performed by Multilocus sequence typing(MLST). OXA-48 was detected in thirteen isolates while three isolates co-expressed OXA-48 and NDM. The mcr-1 gene was absent in all 16 isolates. Deleterious mutations in mgrB included insertion sequences IS903 and ISkpn26 and a premature stop codon. A total of 18 point mutations were identified in PhoPQ and PmrAB TCS; of which, novel mutations were reported in phoQ (K46E, L322V, D152N, F373L, R249G), pmrB (P159R) and pmrA (D149L). Six different sequence types ST231, ST147, ST395, ST42, ST14 and ST101 were identified. Phylogenetic analysis showed that sequence types ST14, ST395 and ST147 are closely related to ST101 and all identified sequence types had a common ancestor ST231. Colistin resistance in K. pneumoniae was attributed to mutations in PhoPQ and PmrAB TCS, while location specific distribution of strains indicates clonal transmission. The results of this study will help in formulation of effective infection prevention and antimicrobial development strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Aghapour Z, Gholizadeh P, Ganbarov K, Bialvaei AZ, Mahmood SS, Tanomand A, et al. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist. 2019;12:965–75.
Kallel H, Bahloul M, Hergafi L, Akrout M, Ketata W, Chelly H, et al. Colistin as a salvage therapy for nosocomial infections caused by multidrug-resistant bacteria in the ICU. Int J Antimicrob Agents. 2006;28:366–9.
Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643.
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8.
Mohapatra DP, Debata NK, Singh SK. Extensively drug-resistant and pandrug-resistant Gram-negative bacteria in a tertiary-care hospital in Eastern India: a 4-year retrospective study. J Glob Antimicrob Resist. 2018;15:246–9.
Pragasam AK, Shankar C, Veeraraghavan B, Biswas I, Nabarro LEB, Inbanathan FY, et al. Molecular mechanisms of colistin resistance in klebsiella pneumoniae causing bacteremia from india—a first report. Front Microbiol. 2017;7:2135.
CLSI Document M100. 27th ed. Wayne, PA: CLSI; 2017. Clinical and laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing.
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.0.2017. [Accessed on 7 Jun 2017]. http://www.eucast.org.
Jaggi N, Chatterjee N, Singh V, Giri S, Dwivedi P, Panwar R, et al. Carbapenem resistance in Escherichia coli and Klebsiella pneumoniae among Indian and international patients in North India. Acta Microbiol Immunol Hungarica. 2019;66:367–76.
Mohan B, Hallur V, Singh G, Sandhu HK, Appannanavar SB, Taneja N. Occurrence of blaNDM-1 & absence of blaKPC genes encoding carbapenem resistance in uropathogens from a tertiary care centre from north India. Indian J Med Res. 2015142:336–43.
Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother. 2014;58:4762–6.
Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–82.
EUCAST. 2016. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. EUCAST http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
Kadri SS, Adjemian J, Lai YL, Spaulding AB, Ricotta E, Prevots DR, et al. National Institutes of Health Antimicrobial Resistance Outcomes Research Initiative (NIH–ARORI), Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin Infect Dis. 2018;67:1803–14.
Taneja N, Singh G, Singh M, Sharma M. Emergence of tigecycline & colistin resistant Acenetobacter Baumanii in patients with complicated UTI in North India. Indian J Med Res. 2011;133:681–4. 6
Lellouche J, Schwartz D, Elmalech N, Ben Dalak MA, Temkin E, Paul M.AIDA study group et al. Combining VITEK® 2 with colistin agar dilution screening assist timely reporting of colistin susceptibility. Clin Microbiol Infect. 2019;25:711–6.
Dafopoulou K, Zarkotou O, Dimitroulia E, et al. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother. 2015;59:4625–30.
Hindler AJ, Humphries RM, Colistin MIC. Variability by method for contemporary clinical isolates of multidrug-resistant gram-negative Bacilli. J Clin Microbiol. 2013;51:1678–84.
Suzuki S, Horinouchi T, Furusawa C. Prediction of antibiotic resistance by gene expression profiles. Nat Commun. 2014;5:5792.
Pitt ME, Elliott AG, Cao MD, et al. Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae. Microb Genom. 2018;4:e000158.
Arena F, Di Pilato V, Vannetti F, et al. Population structure of KPC carbapenemase-producing Klebsiella pneumoniae in a long-term acute-care rehabilitation facility: identification of a new lineage of clonal group 101, associated with local hyperendemicity. Microb Genom. 2020;6:e000308.
Kathayat D, Antony L, Deblais L, Helmy YA, Scaria J, Rajashekara G. Small molecule adjuvants potentiate colistin activity and attenuate resistance development in Escherichia coli by Affecting pmrAB system. Infect Drug Resist. 2020;13:2205–22.
Aires CAM, Pereira PS, Asensi MD, et al. mgrB mutations mediating polymyxin b resistance in klebsiella pneumoniae isolates from rectal surveillance swabs in Brazil. Antimicrob Agents Chemother. 2016;60:6969–72.
Haeili M, Javani A, Moradi J, Jafari Z, Feizabadi MM, Babaei E. MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front Microbiol. 2017;8:2470.
Esposito EP, Cervoni M, Bernardo M, Crivaro V, Cuccurullo S, Imperi F, et al. Molecular epidemiology and virulence profiles of colistin-resistant Klebsiella pneumoniae blood isolates from the hospital agency “Ospedale dei Colli,” Naples, Italy. Front Microbiol. 2018;9:1463.
Shankar C, Mathur P, Venkatesan M, et al. Rapidly disseminating blaOXA-232 carrying Klebsiella pneumoniae belonging to ST231 in India: multiple and varied mobile genetic elements. BMC Microbiol. 2019;19:137.
Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–75.
Cubero M, Cuervo G, Dominguez MÁ, Tubau F, Martí S, et al. Carbapenem-resistant and carbapenem-susceptible isogenic isolates of Klebsiella pneumoniae ST101 causing infection in a tertiary hospital. BMC Microbiol. 2015;15:177.
Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7:1–9.
Roe CC, Vazquez AJ, Esposito EP, Zarrilli R, Sahl JWDiversity. Virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front Microbiol. 2019;10:542.
Acknowledgements
The authors would like to express their gratitude to Prof. Pallab Ray, Post Graduate Institute of Medical Education and Research, Chandigarh, India and Prof. Balaji Veeraraghavan, Department of Clinical Microbiology, Christian Medical College, Vellore, India for providing mcr-1 positive strain and carbapenemases positive strains respectively. Part of these results has also been presented as a poster at the AIIMS ASM-2020 conference in New Delhi, India in October 2020.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. PKN—Study design, Laboratory methods, data analysis and manuscript writing; NC—Study design, Laboratory methods; manuscript correction RP—Isolate collection and Antibiotic susceptibility testing; MD and NJ—Study design and manuscript correction. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics statement and consent for publication
The study protocol was approved by Artemis Hospital Institutional Ethics Committee (AHS-IEC Reg No: ECR/53/Inst/HR/2013/RR-16). Waiver for consent was obtained as microbiological samples were considered a part of standard care procedures and keeping patient identity anonymous in the study. All authors provide their consent for publication.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nirwan, P.K., Chatterjee, N., Panwar, R. et al. Mutations in two component system (PhoPQ and PmrAB) in colistin resistant Klebsiella pneumoniae from North Indian tertiary care hospital. J Antibiot 74, 450–457 (2021). https://doi.org/10.1038/s41429-021-00417-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-021-00417-2
This article is cited by
-
Situation Report on mcr-Carrying Colistin-Resistant Clones of Enterobacterales: A Global Update Through Human-Animal-Environment Interfaces
Current Microbiology (2024)
-
Antimicrobial Resistance Profiles of Multidrug-Resistant Enterobacteria Isolated from Feces of Weaned Piglets
Current Microbiology (2024)