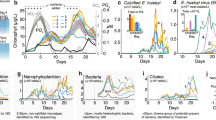
Abstract
Diatom primary productivity is tightly coupled with carbon export through the ballasted nature of the silica-based cell wall, linking the oceanic silicon and carbon cycles. However, despite low productivity, iron (Fe)-limited regimes are considered ‘hot spots’ of diatom silica burial with enhanced carbon export efficiency, raising questions about the mechanisms driving the biogeochemistry of these regions. Marine viruses are classically recognized as catalysts of remineralization through host lysis, short-circuiting the trophic transfer of carbon and facilitating the retention of dissolved organic matter and associated elements in the surface ocean. Here we used metatranscriptomic analysis of diatoms and associated viruses, along with a suite of physiological and geochemical metrics, to study the interaction between diatoms and viruses in Fe-limited regimes of the northeast Pacific. We found low cell-associated diatom virus diversity and abundance in a chronically Fe-limited region of the subarctic northeast Pacific. In a coastal upwelling region of the California Current, transient iron limitation also substantially reduced viral replication. These observations were recapitulated in Fe-limited cultures of the bloom-forming, centric diatom, Chaetoceros tenuissimus, which exhibited delayed virus-mediated mortality in addition to reduced viral replication. We suggest Fe-limited diatoms escape viral lysis and subsequent remineralization in the surface ocean, providing an additional mechanism contributing to enhanced carbon export efficiency and silica burial in Fe-limited oceanic regimes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All cruise-related data are publicly available at the Biological & Chemical Oceanography Data Management Office (CUZ: project number 559966, https://www.bco-dmo.org/deployment/559966; Line P: http://www.waterproperties.ca/linep/2015-009/index.php). Metatranscriptome sequencing data are available in the NCBI sequence read archive (SRA) under the BioProject accession numbers PRJNA320398 and PRJNA388329. Assembled contigs, read counts and functional annotation of contigs are available at http://marchettilab.web.unc.edu/data. All data generated or analysed during this study are included in this published article and its Supplementary Information files. Source data are provided with this paper.
References
Nelson, D. M., Tréguer, P., Brzezinski, M. A., Leynaert, A. & Quéguiner, B. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 9, 359–372 (1995).
Smetacek, V. et al. Deep carbon export from a Southern Ocean iron-fertilized diatom bloom. Nature 487, 313–319 (2012).
Hutchins, D. A., DiTullio, G. R., Zhang, Y. & Bruland, K. W. An iron limitation mosaic in the California upwelling regime. Limnol. Oceanogr. 43, 1037–1054 (1998).
Bruland, K. W., Rue, E. L. & Smith, G. J. Iron and macronutrients in California coastal upwelling regimes: implications for diatom blooms. Limnol. Oceanogr. 46, 1661–1674 (2001).
Boyd, P. W. et al. Mesoscale iron enrichment experiments 1993–2005: synthesis and future directions. Science 315, 612–617 (2007).
Brzezinski, M. A. et al. Enhanced silica ballasting from iron stress sustains carbon export in a frontal zone within the California Current. J. Geophys. Res. Oceans 120, 4654–4669 (2015).
Arteaga, L. A., Pahlow, M., Bushinsky, S. M. & Sarmiento, J. L. Nutrient controls on export production in the Southern Ocean. Glob. Biogeochem. Cycles 33, 942–956 (2019).
Stukel, M. R. & Barbeau, K. A. Investigating the nutrient landscape in a coastal upwelling region and its relationship to the biological carbon pump. Geophys. Res. Lett. 47, e2020GL087351 (2020).
Hutchins, D. A. & Bruland, K. W. Iron-limited diatom growth and Si:N uptake ratios in a coastal upwelling regime. Nature 393, 561–564 (1998).
Takeda, S. Influence of iron availability on nutrient consumption ratio of diatoms in oceanic waters. Nature 393, 774–777 (1998).
Pichevin, L. E., Ganeshram, R. S., Geibert, W., Thunell, R. & Hinton, R. Silica burial enhanced by iron limitation in oceanic upwelling margins. Nat. Geosci. 7, 541–546 (2014).
Brzezinski, M. A. et al. A switch from Si(OH)4 to NO3− depletion in the glacial Southern Ocean. Geophys. Res. Lett. 29, 1564 (2002).
Matsumoto, K., Sarmiento, J. L. & Brzezinski, M. A. Silicic acid leakage from the Southern Ocean: a possible explanation for glacial atmospheric pCO2. Glob. Biogeochem. Cycles 16, 1031 (2002).
Sarmiento, J. L., Gruber, N., Brzezinski, M. A. & Dunne, J. P. High-latitude controls of thermocline nutrients and low latitude biological productivity. Nature 427, 56–60 (2004).
Fuhrman, J. A. Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999).
Suttle, C. A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Wilhelm, S. W. & Suttle, C. A. Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49, 781–788 (1999).
Kranzler, C. F. et al. Silicon limitation facilitates virus infection and mortality of marine diatoms. Nat. Microbiol. 4, 1790–1797 (2019).
Laber, C. P. et al. Coccolithovirus facilitation of carbon export in the North Atlantic. Nat. Microbiol. 3, 537–547 (2018).
Yamada, Y., Tomaru, Y., Fukuda, H. & Nagata, T. Aggregate formation during the viral lysis of a marine diatom. Front. Mar. Sci. 5, 167 (2018).
Pelusi, A. et al. Virus-induced spore formation as a defense mechanism in marine diatoms. New Phytol. 229, 2251–2259 (2020).
Johnson, K. S., Chavez, F. P. & Friederich, G. E. Continental-shelf sediment as a primary source of iron for coastal phytoplankton. Nature 398, 697–700 (1999).
Harrison, P. J. Station Papa time series: insights into ecosystem dynamics. J. Oceanogr. 58, 259–264 (2002).
Marchetti, A. et al. Development of a molecular-based index for assessing iron status in bloom-forming pennate diatoms. J. Phycol. 53, 820–832 (2017).
Cohen, N. R. et al. Diatom transcriptional and physiological responses to changes in iron bioavailability across ocean provinces. Front. Mar. Sci. 4, 360 (2017).
Lampe, R. H. et al. Different iron storage strategies among bloom-forming diatoms. Proc. Natl Acad. Sci. USA 115, E12275–E12284 (2018).
King, A. L. & Barbeau, K. Evidence for phytoplankton iron limitation in the southern California Current System. Mar. Ecol. Prog. Ser. 342, 91–103 (2007).
Boyd, P. & Harrison, P. J. Phytoplankton dynamics in the NE subarctic Pacific. Deep Sea Res. II 46, 2405–2432 (1999).
Till, C. P. et al. The iron limitation mosaic in the California Current System: factors governing Fe availability in the shelf/near-shelf region. Limnol. Oceanogr. 64, 109–123 (2019).
Gozzelino, R., Jeney, V. & Soares, M. P. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 50, 323–354 (2010).
Richaud, C. & Zabulon, G. The heme oxygenase gene (pbsA) in the red alga Rhodella violacea is discontinuous and transcriptionally activated during iron limitation. Proc. Natl Acad. Sci. USA 94, 11736–11741 (1997).
Allen, A. E. et al. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl Acad. Sci. USA 105, 10438–10443 (2008).
Thamatrakoln, K., Korenovska, O., Niheu, A. K. & Bidle, K. D. Whole-genome expression analysis reveals a role for death-related genes in stress acclimation of the diatom Thalassiosira pseudonana. Environ. Microbiol. 14, 67–81 (2012).
Marchetti, A. et al. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl Acad. Sci. USA 109, E317–E325 (2012).
De La Rocha, C. L., Hutchins, D. A., Brzezinski, M. A. & Zhang, Y. Effects of iron and zinc deficiency on elemental composition and silica production by diatoms. Mar. Ecol. Prog. Ser. 195, 71–79 (2000).
Leynaert, A. et al. Effect of iron deficiency on diatom cell size and silicic acid uptake kinetics. Limnol. Oceanogr. 49, 1134–1143 (2004).
van Creveld, S. G., Rosenwasser, S., Levin, Y. & Vardi, A. Chronic iron limitation confers transient resistance to oxidative stress in marine diatoms. Plant Physiol. 172, 968–979 (2016).
Slagter, H. A., Gerringa, L. J. A. & Brussaard, C. P. D. Phytoplankton virus production negatively affected by iron limitation. Front. Mar. Sci. 3, 156 (2016).
Drakesmith, H. & Prentice, A. Viral infection and iron metabolism. Nat. Rev. Microbiol. 6, 541–552 (2008).
Weinbauer, M. G., Arrieta, J. M., Griebler, C. & Herndlb, G. J. Enhanced viral production and infection of bacterioplankton during an iron-induced phytoplankton bloom in the Southern Ocean. Limnol. Oceanogr. 54, 774–784 (2009).
Torres, M. A., Jones, J. D. G. & Dangl, J. L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378 (2006).
Sheyn, U., Rosenwasser, S., Ben-Dor, S., Porat, Z. & Vardi, A. Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. ISME J. 10, 1742–1754 (2016).
Hyodo, K., Hashimoto, K., Kuchitsu, K., Suzuki, N. & Okuno, T. Harnessing host ROS-generating machinery for the robust genome replication of a plant RNA virus. Proc. Natl Acad. Sci. USA 114, E1282–E1290 (2017).
Espinoza, J. A., Gonzalez, P. A. & Kalergis, A. M. Modulation of antiviral immunity by heme oxygenase-1. Am. J. Pathol. 187, 487–493 (2017).
Durkin, C. A. et al. Frustule-related gene transcription and the influence of diatom community composition on silica precipitation in an iron-limited environment. Limnol. Oceanogr. 57, 1619–1633 (2012).
Assmy, P. et al. Thick-shelled, grazer-protected diatoms decouple ocean carbon and silicon cycles in the iron-limited Antarctic Circumpolar Current. Proc. Natl Acad. Sci. USA 110, 20633–20638 (2013).
Kimura, K. & Tomaru, Y. Effects of temperature and salinity on diatom cell lysis by DNA and RNA viruses. Aquat. Microb. Ecol. 79, 79–83 (2017).
Thamatrakoln, K. et al. Light regulation of coccolithophore host–virus interactions. New Phytol. 221, 1289–1302 (2019).
Zimmerman, A. E. et al. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat. Rev. Microbiol. 18, 21–34 (2020).
Brzezinski, M. A. et al. Co-limitation of diatoms by iron and silicic acid in the equatorial Pacific. Deep Sea Res. II 58, 493–511 (2011).
Boyer, T. P. et al. World Ocean Database 2013 (NOAA Atlas, 2013).
Krause, J. W. et al. The interaction of physical and biological factors drives phytoplankton spatial distribution in the northern California Current. Limnol. Oceanogr. 65, 1974–1989 (2020).
Krause, J. W., Nelson, D. M. & Brzezinski, M. A. Biogenic silica production and the diatom contribution to primary production and nitrate uptake in the eastern equatorial Pacific Ocean. Deep Sea Res. II 58, 434–448 (2011).
Brzezinski, M. A. & Phillips, D. R. Evaluation of 32Si as a tracer for measuring silica production rates in marine waters. Limnol. Oceanogr. 42, 856–865 (1997).
Nelson, D. M., Brzezinski, M. A., Sigmon, D. E. & Franck, V. M. A seasonal progression of Si limitation in the Pacific sector of the Southern Ocean. Deep Sea Res. II 48, 3973–3995 (2001).
Krause, J. W., Brzezinski, M. A., Villareal, T. A. & Wilson, C. Increased kinetic efficiency for silicic acid uptake as a driver of summer diatom blooms in the North Pacific subtropical gyre. Limnol. Oceanogr. 57, 1084–1098 (2012).
Birol, I. et al. De novo transcriptome assembly with ABySS. Bioinformatics 25, 2872–2877 (2009).
Robertson, G. et al. De novo assembly and analysis of RNA-seq data. Nat. Methods 7, 909–912 (2010).
Gremme, G., Steinbiss, S. & Kurtz, S. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans. Comput. Biol. Bioinform. 10, 645–656 (2013).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Keeling, P. J. et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12, e1001889 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Wagner, G. P., Kin, K. & Lynch, V. J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 (2012).
Alexander, H., Jenkins, B. D., Rynearson, T. A. & Dyhrman, S. T. Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc. Natl Acad. Sci. USA 112, E2182–E2190 (2015).
Lampe, R. H. et al. Divergent gene expression among phytoplankton taxa in response to upwelling. Environ. Microbiol. 20, 3069–3082 (2018).
Warnes, G. R. et al. gplots: Various R Programming Tools for Plotting Data https://cran.r-project.org/web/packages/gplots/index.html (2019).
Oksanen, J. et al. vegan: Community Ecology Package https://cran.r-project.org/web/packages/vegan/index.html (2019).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Matsen, F. A., Kodner, R. B. & Armbrust, E. V. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinform. 11, 538 (2010).
Shirai, Y. et al. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl. Environ. Microbiol. 74, 4022–4027 (2008).
Chen, L.-M., Edelstein, T. & McLachlan, J. Bonnemaisonia hamifera Hariot in nature and in culture. J. Phycol. 5, 211–220 (1969).
Harrison, P. J., Waters, R. E. & Taylor, F. J. R. A broad spectrum artificial sea water medium for coastal and open ocean phytoplankton. J. Phycol. 16, 28–35 (1980).
Berges, J. A., Franklin, D. J. & Harrison, P. J. Evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the last two decades. J. Phycol. 37, 1138–1145 (2001).
Sunda, W. G., Price, N. M. & Morel, F. M. M. Trace metal ion buffers and their use in culture studies. Algal Cult. Tech. 4, 35–63 (2005).
Tomaru, Y., Shirai, Y., Toyoda, K. & Nagasaki, K. Isolation and characterization of a single-stranded DNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus. Aquat. Microb. Ecol. 64, 175–184 (2011).
Parsons, T. R. A Manual of Chemical & Biological Methods for Seawater Analysis (Elsevier, 2013).
Krause, J. W., Lomas, M. W. & Nelson, D. M. Biogenic silica at the Bermuda Atlantic time-series study site in the Sargasso Sea: temporal changes and their inferred controls based on a 15-year record. Glob. Biogeochem. Cycles 23, GB3004 (2009).
Gorbunov, M. Y. & Falkowski, P. G. Fluorescence induction and relaxation (FIRe) technique and instrumentation for monitoring photosynthetic processes and primary production in aquatic ecosystems. In Photosynthesis: Fundamental Aspects to Global Perspectives—Proc. 13th International Congress of Photosynthesis (eds Van der Est, A. & Bruce, D.) 1029–1031 (Allen and Unwin, 2004).
Suttle, C. A. in Handbook of Methods in Aquatic Microbial Ecology (eds Kemp, P. F. et al.) 121–134 (CRC Press, 1993).
Klee, A. J. A computer program for the determination of most probable number and its confidence limits. J. Microbiol. Methods 18, 91–98 (1993).
Acknowledgements
We would like to thank the captain and the crew of the RV Melville (MV1405) and the Canadian Coast Guard Ship J. P. Tully (Line P 2015-09), as well as G. Smith, F. Kuzminov, K. Ellis and T. Coale for technical assistance during the cruise. Thank you to Y. Tomaru for providing the laboratory diatom host-virus systems, K. Bondoc and B. Knowles for useful discussions on statistical analyses and K. D. Bidle for thoughtful feedback on the manuscript. This work was supported by grants from the National Science Foundation (OCE-1333929 to K.T., OCE-1334387 to M.A.B., OCE-1334935 to A.M., OCE-1259776 to K.W.B. and OCE-1334632 to B.S.T.) and a postdoctoral fellowship to C.F.K. from the Simons Foundation (SF 548156). Salary support for C.F.K. was also provided by the Institute of Earth, Ocean and Atmospheric Sciences at Rutgers University, the Rappaport Fund for Advanced Studies and Israel’s Council for Higher Education.
Author information
Authors and Affiliations
Contributions
C.F.K. and K.T. conceived the project, designed the laboratory experiments and wrote the manuscript. C.F.K. performed metatranscriptome and statistical analyses. C.F.K. conducted the laboratory culture-based experiments. J.M. and J.R.L. provided technical support on laboratory experiments. M.M. assisted with RdRp phylogenetic analyses. K.T., M.A.B., N.R.C., M.M., C.P.T., A.M., B.S.T. and K.W.B. participated, collected and analysed samples on the CUZ cruise; B.S.T. and N.R.C. participated, collected and analysed samples on the Line P cruise. A.M., N.R.C. and R.H.L. extracted the RNA and generated the metatranscriptome data. All authors provided comments on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Geoscience thanks Erin Bertrand, Alex Poulton and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Clare Davis; Xujia Jiang.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Bulk parameters measured during incubations at each site.
a, Size-fractionated chlorophyll a (>5 µm; μg L−1), b, biogenic silica (μM), and c, maximum photochemical quantum yield of photosystem II (photosynthetic efficiency; Fv/Fm) throughout each incubation. Treatments included an initial sample (t0; open), and unamended Control (Ctrl; yellow), DFB (blue) and +Fe (orange). d, Fold-change in chlorophyll a (open bars) and biogenic silica (filled bars) between +Fe and Ctrl treatments at each timepoint. Note that at sites S2 and P4, 15 μM of Si(OH)4 and 10 µM of NO3− were added, respectively, to all treatments due to low initial concentrations (Si(OH)4 at S2 < 4.7 μM; NO3− at P4 < 1.5 μM). Mean and standard error of triplicate incubations are shown along with individual replicates (diamonds). Statistical significance of the community response to iron addition is depicted (panels a-c) for each time point with ***P < 0.001, **P < 0.01, *P < 0.05 by analysis of variance (ANOVA) followed by Tukey’s HSD post hoc test. Additional statistical analysis is available in Supplementary Data 2.
Extended Data Fig. 2 Silicon stress during incubation experiments at each site.
Kinetic limitation of Si uptake, or Si stress (Vamb:Venh), at each site for initial samples (t0; open diamonds) and unamended Control (Ctrl; yellow), DFB (blue) and +Fe (orange) treatments throughout each incubation experiment. Each point represents a distinct treatment (color) and time point (symbol) within each incubation (diamonds, t0; circles, t1, 24–48 h; triangles, t2, 48–96 h). See methods and Supplementary Fig. 1 for individual time points. Values approaching zero are indicative of severe Si stress and values ~1 are indicative of silicon replete populations. The boxes depict the median (horizontal line) and upper and lower quartiles of the data with whiskers encompassing data points within 1.5× of the interquartile range. ***P < 0.001 by Kruskal-Wallis with Dunn’s multiple comparison test. No Si stress data was collected during the P4 and P26 incubations, or at t2 in the S1 and S2 incubations for Ctrl and +Fe treatments.
Extended Data Fig. 3 Phylogenetic analysis and size distribution of diatom virus-like contigs.
a, Maximum likelihood phylogenetic tree of RNA-dependent RNA polymerase (RdRp) amino acid sequences within the order Picornavirales and placement of homologous contigs identified in metatranscriptomes. Bootstrap values >50 are shown (100 replicates). Triangles denote sequences that fall within the family Marnaviridae with blue triangles denoting the genus Marnavirus, which is distinctly comprised of dinoflagellate viruses and red triangles identifying putative diatom viruses that were selected for downstream analysis. b, Size distribution (bp) of diatom virus-like contigs. The abbreviations, names and NCBI database accession numbers of the amino acid sequences used to construct the reference alignment are: AglaRNAV, Asterionellopsis glacialis RNA virus, BAP16719; CsfrRNAV, Chaetoceros socialis radians RNA virus 1, YP_002647032; RsRNAV, Rhizosolenia setigera RNA virus 01, YP_006732323; CtenRNAV01, C. tenuissimus RNA virus 01, YP_009505620; CtenRNAVII, C. tenuissimus RNA virus type-II, BAP99818; CspRNAV2, Chaetoceros species RNA virus 02, BAK40203; HaRNAV, Heterosigma akashiwo RNA virus, AAP97137; CPSMV, cowpea severe mosaic virus, NP_619518; BPMV, Bean pod mottle virus, NP_612349; PYFV, Parsnip yellow fleck virus, BAA03151; RTSV, Rice tungro spherical virus, NP_042507;SBV, Sacbrood virus, AIZ75645; PV, Human poliovirus 1, CAA24461; AIV, Aichi virus 1, ADN52312; BQCV, Black queen cell virus, NP_620564; TrV, Triatoma virus, NP_620562; CPV, Cricket paralysis virus, NP_647481; DCV, Drosophila C virus, NP_044945; TSV, Taura syndrome virus, NP_149057.
Extended Data Fig. 4 Temporal changes in cell-associated diatom viruses throughout each incubation.
Log2 fold change in diatom virus contig abundance (nCv) at t1 (circles, 24–48 h) or t2 (triangles, when present, 48–72 h) compared to t0 for Ctrl (yellow), DFB (blue) and +Fe (orange) treatments (n = 3), *P < 0.05 by a one-way analysis of variance (ANOVA) followed by a Tukey HSD post hoc test. The boxes depict the median (horizontal line) and upper and lower quartiles of the data with whiskers encompassing data points within 1.5× of the interquartile range. See methods and Supplementary Fig. 1 for individual time points (symbols) in each incubation.
Extended Data Fig. 5 Temporal changes in cell-associated diatom viruses throughout the S2 incubation.
a, Log2 fold change in diatom virus contig abundance (nCv) between t48 and t0 for Ctrl (yellow), DFB (blue) and +Fe (orange) treatments (n = 3), *P < 0.05 by a one-way analysis of variance (ANOVA) followed by a Tukey HSD post hoc test. The boxes depict the median (horizontal line) and upper and lower quartiles of the data with whiskers encompassing data points within 1.5× of the interquartile range. b, Heatmap of mean abundance for each diatom virus contig (nCv) identified in the initial (t0) sample and at t48 across the treatments for both high (top panel) and low abundance (bottom panel) contigs. Contigs that were below detection are depicted in white. c, Abundance (nCv) of each cell-associated diatom virus contig in Ctrl (left panel), DFB (middle panel) and +Fe (right panel) treatments at t48 vs t0. Mean ± standard error is shown (n = 3). Dotted lines denote unity-slope lines, indicating no difference between timepoints. Insets depict ‘high’ abundance contigs. In panels a and c, open and closed symbols denote ‘high’ and ‘low’ abundance diatom virus contigs, respectively.
Extended Data Fig. 6 Diagnosing iron limitation in Chaetoceros tenuissimus.
a, Specific growth rates (day−1), b, mean chlorophyll autofluorescence (RFU cell−1) and c, electron transport rates (e− s−1 PSII−1) of C. tenuissimus cultures in replete (orange symbols) and Fe-limited (blue symbols) growth media. d, Cell abundance and e, maximum photochemical quantum yield of photosystem II (photosynthetic efficiency; Fv/Fm) during Fe-limitation (blue) and after Fe addition (orange) in an Fe ‘rescue’ experiment. Each independent biological replicate (n = 3) is shown by individual symbols with lines of best fit depicting a LOESS regression. f, cellular biogenic silica (µmol bSiO2 cell−1) and g, cellular antioxidant capacity (mM Trolox eq cell−1) of replete and iron-limited C. tenuissimus. Statistical significance was determined using an unpaired, two-sided t-test. Different symbols denote biologically independent replicates across six independent experiments.
Extended Data Fig. 7 The impact of Fe limitation on diatom host-virus dynamics in laboratory cultures of Chaetoceros tenuissimus infected with CtenRNAV.
Host abundance in replete (orange circles) and Fe-limited (blue triangles), uninfected (open symbols) and infected (closed symbols) cultures with the single stranded (ss) RNA-containing virus, CtenRNAV. Individual symbols represent independent biological replicates (n = 3) with lines of best fit depicting a LOESS regression.
Extended Data Fig. 8 A role for oxidative stress and reactive oxygen species during infection of Chaetoceros tenuissimus.
a, Host abundance and b, intracellular levels of reactive oxygen species (ROS; assessed by the diagnostic, fluorescent stain H2DCFDA in replete, uninfected (open, orange symbols/bars) and infected (closed, orange symbols/bars) C. tenuissimus cultures with CtenDNAV. Mean ± standard error is shown (n = 3). c, Host abundance and d, maximum photochemical quantum yield of photosystem II (photosynthetic efficiency; Fv/Fm) following addition of different doses of hydrogen peroxide (H2O2; 0-150 µM). Mean ± standard error is shown for biological duplicates. Due to the observed decrease in cell abundance and photosynthetic efficiency, followed by physiological recovery, 100 µM H2O2 was chosen for downstream infection experiments. Time course of e, host abundance and f, photosynthetic efficiency during viral infection with CtenDNAV of untreated control cultures (orange circles) and cultures pre-exposed to a sub- lethal dose of H2O2 (100 µM; blue squares) for uninfected (open symbols) and infected (closed symbols) cultures (n = 3). Data are representative of three independent experiments.
Supplementary information
Supplementary Data 1
Data 1a Taxonomic assignment of mapped reads: taxa. Data 1b Taxonomic assignment of mapped reads: diatom genera. Data 2 Summary of statistical analyses. Data 3a Diatom virus TBLASTN hits. Data 3b Putative diatom virus contig abundance. Data 4a Differentially expressed contigs_S2_DFB/Fe. Data 4b Differentially expressed contigs_S2_Fe/Ctrl. Data 4c Differentially expressed contigs_S2_DFB/Ctrl.
Source data
Source Data Fig. 1
Numerical data.
Source Data Fig. 2
Numerical data.
Source Data Fig. 3
Numerical data, Supplementary Data 3 and 4.
Source Data Fig. 4
Numerical data.
Source Data Extended Data Fig. 1
Numerical data.
Source Data Extended Data Fig. 2
Numerical data.
Source Data Extended Data Fig. 6
Numerical Data.
Source Data Extended Data Fig. 7
Numerical Data.
Source Data Extended Data Fig. 8
Numerical Data.
Rights and permissions
About this article
Cite this article
Kranzler, C.F., Brzezinski, M.A., Cohen, N.R. et al. Impaired viral infection and reduced mortality of diatoms in iron-limited oceanic regions. Nat. Geosci. 14, 231–237 (2021). https://doi.org/10.1038/s41561-021-00711-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41561-021-00711-6
This article is cited by
-
Establishment of a simple method for cryopreservation of the marine diatoms, Chaetoceros and Phaeodactylum
Journal of Applied Phycology (2023)
-
Reduced bacterial mortality and enhanced viral productivity during sinking in the ocean
The ISME Journal (2022)
-
Diminished carbon and nitrate assimilation drive changes in diatom elemental stoichiometry independent of silicification in an iron-limited assemblage
ISME Communications (2022)
-
Shunt or shuttle
Nature Geoscience (2021)