Abstract
Endophytes are microbes that live, for at least a portion of their life history, within plant tissues. Endophyte assemblages are often composed of a few abundant taxa and many infrequently observed, low-biomass taxa that are, in a word, rare. The ways in which most endophytes affect host phenotype are unknown; however, certain dominant endophytes can influence plants in ecologically meaningful ways—including by affecting growth and immune system functioning. In contrast, the effects of rare endophytes on their hosts have been unexplored, including how rare endophytes might interact with abundant endophytes to shape plant phenotype. Here, we manipulate both the suite of rare foliar endophytes (including both fungi and bacteria) and Alternaria fulva–a vertically transmitted and usually abundant fungus–within the fabaceous forb Astragalus lentiginosus. We report that rare, low-biomass endophytes affected host size and foliar %N, but only when the heritable fungal endophyte (A. fulva) was not present. A. fulva also reduced plant size and %N, but these deleterious effects on the host could be offset by a negative association we observed between this heritable fungus and a foliar pathogen. These results demonstrate how interactions among endophytic taxa determine the net effects on host plants and suggest that the myriad rare endophytes within plant leaves may be more than a collection of uninfluential, commensal organisms, but instead have meaningful ecological roles.
Similar content being viewed by others
Introduction
Plants are intimately associated with numerous fungi and bacteria that live within leaves, roots, stems, and other tissues [1, 2]. These microbes, termed endophytes [3] are ubiquitous and occur in hosts representing all major lineages of plants [4]. Over the last 20 years, it has become clear that dominant endophytic taxa can have dramatic ecological consequences–a finding demonstrated particularly well in studies manipulating the abundance of vertically transmitted fungi occurring within cool-season, perennial grasses [5, 6]. For example, these fungi can influence successional trajectories of vegetation [7, 8], reshape host-associated arthropod assemblages [9], and mediate host reproductive output [10]. In contrast, the ecological roles of rare endophytes—which we define as those taxa that are infrequently encountered and of low biomass—remain largely unexamined, despite that fact that these rare taxa constitute the bulk of biodiversity present within endophyte assemblages. Here, we manipulate both rare and dominant endophytes living within a perennial forb to characterize how these taxa interact and affect host phenotype.
Most endophytes are horizontally transmitted among mature hosts via rainfall, air currents, or arthropods [11, 12] and colonize only a few cubic millimeters of host tissue [13]. Given the low biomass of these rare taxa, it is tempting to downplay their importance. However, examples from macroorganism community ecology demonstrate that certain “keystone” species, despite relatively low abundance, can exert community-wide influence [14]. For instance, beavers are uncommon mammals, yet, by reshaping fluvial geomorphology, they have profound influence on co-occurring aquatic animals, waterfowl, and riparian plants [15]. Similarly, rare endophytes could function as keystone species via several mechanisms, including by influencing the host phenotype, catabolism of low-concentration compounds into products required by other microbial taxa, or synthesis of potent bioactive compounds [16,17,18].
However, the ecological influence of rare endophytes need not be the purview of just a few species. Instead, minor effects of individual taxa could accrue to the point of assemblage-wide relevance—just as numerous genetic variants, each of minimal influence, commonly underlie phenotypes [19]. For example, an individual endophytic bacterium may trigger a highly localized immune response of negligible importance for the host and co-occurring endophytes. But the combined effects of many bacteria might initiate systemic acquired resistance within plants, with important implications for pathogen resistance and endophyte community assembly [20, 21].
Ascribing ecological influence to endophytic taxa, rare or otherwise, is complicated by a lack of understanding regarding how endophytes mediate plant trait expression [22, 23]. While the effects of certain endophytes on host growth promotion [24] and pathogen resistance [25,26,27] have attracted attention, few studies have examined endophyte mediation of other traits—including, for example, functional traits such as specific leaf area (e.g. [28]), phenology [29], and foliar elemental concentration [30] (for more, see reviews by [22, 23, 31]). Nevertheless, the handful of studies demonstrating plant trait mediation by endophytes are impressive. For instance, Mejía et al. [32] reported that inoculation of Theobroma cacao trees with the widespread, horizontally transmitted, fungal endophyte Colletotrichum tropicale affected expression of hundreds of host genes, including upregulation of some involved in the ethylene-driven immune response. These authors also found that inoculation decreased photosynthetic rate, increased leaf cellulose and lignin content, and shifted foliar isotopic ratios of nitrogen (N) and carbon (C). Similarly impressive results were reported by Dupont et al. [33] who found colonization of the grass Lolium perenne by the Epichloë festucae endophyte affected transcription of one third of host genes (for slightly more tempered, results see [34]). These studies demonstrate the importance of systemic, or otherwise abundant, endophytes on their hosts, but we are unaware of any studies that manipulate the presence of low biomass, non-systemic endophytes to determine the extent to which they have similar effects on host phenotype.
Here, we perturb the microbial consortium within the fabaceaous forb Astragalus lentiginosus (spotted locoweed) to understand how endophytes belonging to different abundance categories affect plant trait expression. A. lentiginosus is a widespread, perennial forb that grows throughout the arid regions of the western United States of America [35]. A. lentiginosus exhibits extreme phenotypic variation and has over 40 varietal designations [35], making it the most taxonomically rich plant species in North America [36]. A dominant fungal endophyte present within A. lentiginosus is Alternaria fulva (Ascomycota: Dothideomycetes: Pleosporaceae: Alternaria section Undifilum [37,38,39]). A. fulva is a seed-borne endophyte that grows systemically through its host and synthesizes the bioactive alkaloid swainsonine [40]. Consumption of swainsonine-laced tissues by mammalian herbivores can lead to extreme toxicosis and even death [41]. A. fulva is prevalent throughout the range of its host, though not all populations of A. lentiginosus are colonized by the fungus, and intrapopulation variation in fungal colonization has also been reported [42].
Alternaria section Undifilum fungi have been observed in numerous swainsonine-containing taxa within Astragalus and Oxytropis that are colloquially called “locoweeds” [39, 43,44,45]. The nature of the relationship between locoweeds and their seed-borne fungi is somewhat unclear. Swainsonine does not seem to influence certain specialist arthropod herbivores [46, 47], which is suggestive of commensalism [48]. However, recent work supports a more mutualistic relationship between plant and fungus. For instance, Harrison et al. [42] demonstrated, via a DNA sequence-based survey, that swainsonine concentrations and A. fulva relative abundance were inversely related to fungal endophyte richness, potentially reducing exposure of hosts to pathogens. In a culture-based survey, Lu et al. [49] reported similar results in two other locoweed species (also see [50] for an analogous phenomenon in a grass-Neotyphodium endophyte system). The results from these surveys suggest that vertically transmitted Alternaria endophytes can shape fungal endophyte assemblages, though effects on bacterial endophytes are unknown. In addition, Cook et al. [51] demonstrated that Alternaria section Undifilum endophytes can affect the biomass and protein content of several locoweed taxa, including A. lentiginosus. These results suggest A. fulva may mediate other host traits as well.
By removing embryos from the seed coat, the abundance of A. fulva in plant tissues can be greatly reduced or eliminated [52]. We used this approach to manipulate the abundance of A. fulva in A. lentiginosus plants to experimentally test the aforementioned antagonistic relationship between A. fulva and co-occurring endophytes and explore how A. fulva affects various host traits, including size, leaflet area, specific leaf area, foliar C and N, phenology, and nitrogen fixation in the rhizosphere. For a subset of focal plants, we applied an inoculum slurry to leaf surfaces to boost exposure to rare, horizontally transmitted endophytes. We applied these manipulations in a full factorial design to compare how endophytes of differing abundance categories shape host plant phenotype and explore how a dominant endophyte might influence co-occurring fungi and bacteria.
Methods
Field experiment
During the early spring of 2017, seeds of A. lentiginosus var. wahweapensis from the Henry Mts. in Utah, USA (collected in 2005 from a population known to possess A. fulva) were lightly scarified, left to imbibe deionized water overnight and then germinated indoors in a mix of humus, compost, and topsoil sourced from the Reno, NV region. To reduce the relative abundance of the vertically transmitted fungal endophyte, A. fulva embryos were excised from a subset of seeds prior to planting, as per [52]. Notably, this treatment can eliminate the presence of A. fulva from plants altogether, but occasionally only reduces A. fulva abundance. Seedlings were grown at ambient temperature under a 16:8 (light:dark) daily lighting regime and watered with dechlorinated tap water. Individuals from different treatments were interspersed haphazardly and not allowed to touch one another. Seedlings were periodically reorganized to avoid any influence of subtly differing conditions across the growth area. To speed growth, Miracle-Gro (Scotts Miracle-Gro Company, Marysville, OH) was applied several times to all replicates during the first month of growth. To control for possible confounding effects of seed coat removal, embryos were excised from a subset of seeds and planted along with potato dextrose agar (PDA) that was sterile, or that was colonized by A. fulva, which had been cultured from intact seeds. These control seedlings were planted several weeks later than other seedlings, due to slow growth of A. fulva cultures.
In early June, seedlings were installed in five gallon pots filled with equal parts locally sourced humus and topsoil and placed in an abandoned, largely denuded field near the University of Nevada, Reno. No Astragalus taxa were observed growing within this field and the plants that were present in the field were mostly non-native forbs. A. lentiginosus grows in a wide variety of settings, including roadsides and disturbed areas not unlike our site. A total of 300 plants were installed (between 54 and 68 per treatment group, see Table 1). Pots were organized randomly with respect to treatment and were placed one meter apart so plants never touched one another. Dechlorinated water was applied as needed to all plants at the same time (typically every other day, except during the heat of summer when watering was conducted daily). Every 2 weeks a slurry of microbial inocula (described below) was sprayed on leaves of half of the plants. A solution with identical surfactant, but no microbial inoculum, was applied to untreated plants. Plants were left in the field from early June through mid-September, at which point leaves were removed for sequencing and culturing.
Inoculum preparation
Twenty morphologically unique, reproductive fungal isolates were obtained from the following woody shrubs growing near Reno, NV: Artemisia tridentata, Ericameria nauseosa, Prunus andersonii, and Tetradymia canescens. These shrubs are abundant throughout the Great Basin Desert and, consequently, we reasoned they contained horizontally transmitted, foliar microbes likely to be regularly encountered by A. lentiginosus. Indeed, sequencing revealed that most of the fungal taxa within the inoculum were observed in the wild-collected A. lentiginosus individuals examined in [42] (for details see the Supplemental Material). Individual shrubs to be sampled were selected haphazardly. We did not use leaves from Astragalus species to avoid inoculating plants with A. fulva and thus obviating our treatment to reduce this fungus. Leaves were cut into sections (of several mm2) and placed on PDA and the resulting microbial growth isolated and subcultured over 2 weeks. Spores from isolates were removed and suspended in deionized water and 0.0001% TWEEN 20 (Sigma-Aldrich), a detergent that functioned as a surfactant. A haemocytometer was used to dilute the suspension to ~100,000 spores ml−1. This concentration was chosen because it produced no obvious negative symptoms in A. lentiginosus seedlings during preliminary experiments. Aseptic technique was used throughout culturing and inoculum preparation. Two aliqouts of inoculum were sequenced to identify the constituent microbial taxa. We specifically targeted fungi for culturing, but sequencing revealed that bacteria were also present within the inoculum.
Plant trait measurement
All plant traits were measured concomitant with sample collection for foliar microbiome characterization. Plant size was measured as the minimum size box that would enclose the plant. This was calculated as the product of the width of the plant at its widest point, the width of the plant perpendicular to that point, and plant height. Phenological state and number of leaves were characterized for each plant. Area and specific leaf area (SLA; leaflet area divided by mass of leaflet) were measured for three dried leaflets per plant and averaged. Two healthy leaflets were removed from 8–12 leaves per plant, rinsed with tap water, dried in a laminar flow hood (<12 h total) and frozen until further processing. These leaflets were then parsed for microbiome characterization, swainsonine quantification, and carbon (C) and nitrogen (N) analysis. Swainsonine concentration in ~50 mg of dried, ground foliar tissue was measured using an LC–MS/MS approach described in [53]. Briefly, an 18 h extraction in 2% acetic acid with agitation was followed by centrifugation. Supernatant was added to 20 mM ammonium acetate and subjected to LC–MS/MS analysis. Percent C and N and 14 N: 15N isotopic ratios in 3–4 mg dried foliar tissue, were measured by the Nevada Stable Isotope Laboratory using a Micromass Isoprime stable isotope ratio mass spectrometer (Elementar, Stockport, UK) and a Eurovector elemental analyzer (Eurovector, Pavia, Italy). The percentage of nitrogen in tissues due to fixation alone (NDFA) was calculated as per [54] through comparison with samples from co-occurring Chenopodium album, which is not known to harbor nitrogen fixing rhizosphere bacteria.
Sequence and culture-based characterization of the foliar microbiome
We characterized endophytic assemblages through both culturing and DNA sequencing, thus affording us insight into the effects of treatment on microbial assemblages via two complimentary measurement techniques. For our culture-based assay, we choose three leaflets per plant. Leaflets were surface sterilized, cut into 3–4 pieces, and plated onto PDA using aseptic technique. Surface sterilization involved rinsing in 95% ethanol for 30 s, followed by 2 min in 10% sodium hypochlorite solution, 2 min in 70% in ethanol, and a final rinse with deionized water. Preliminary experiments confirmed the success of this surface sterilization technique. Cultures were grown in the dark at ambient temperatures for 1.5 months. Microbial growth (either fungal or bacterial) was isolated, subcultured, and the number of morphologically unique cultures and the percentage of leaf pieces colonized recorded. Cultures corresponding to A. fulva were identified visually through comparison to A. fulva cultures grown from seeds used for this experiment and through sequencing (we did not sequence the other cultures for logistical reasons).
DNA was extracted from three surface-sterilized, dried, and ground leaflets per plant using DNeasy plant mini kits (Qiagen, Hilden, Germany). Extraction blanks for each kit were used as negative controls. Dual-indexed libraries were made at the University of Wyoming and were sequenced on the Illumina NovaSeq platform (paired-end 2 × 250; San Diego, CA, USA) by Psomagen, Inc. (Rockville, MD, USA). To characterize bacterial assemblages, the 16S (V4) locus was amplified using the 515–806 primer pair [55], while for fungal assemblages the ITS1 locus was amplified using the ITS1f-ITS2 primer pair [56]. A synthetic DNA internal standard (ISD) was added to template DNA prior to library creation [57]. In addition, unique synthetic DNAs of our own design were added to each sample to allow cross-contamination to be detected (sensu [58]; we refer to these oligos as “coligos,” which is short for cross-contamination checking oligos). A mock community consisting of eight bacteria and two fungi was also sequenced (Zymo Research, Irvine, CA, USA) as a positive control. PCR was performed in duplicate and unique index sequences were ligated onto each PCR replicate, thus allowing us to determine technical variation due to PCR. For full library preparation details see the Supplemental Material.
Sequence data were demultiplexed using a custom perl script that used Levenshtein distances to correct errors in index sequences and assign reads to samples. Primers and Illumina adapters were removed using cutadapt v1.13-py27 (Martin 2011) and poly-G tails removed using fastp v0.21.0 (poly-G tails occurred due to a lack of signal for very short-template molecules, such as our coligos [see above]);. Paired-end reads were merged using vsearch v2.9.0 [59, 60] with staggering allowed, a minimum of ten overlapping bases, and a maximum of 12 mismatches in the overlapping region. The probability of base-calling errors within merged reads was estimated and those reads expected to include more than one error were discarded.
Using vsearch, unique reads were identified and clustered into OTUs (operational taxonomic units) using the ‘cluster unoise’ algorithm [61] with a minimum of 12 sequences required, each with a minimum length of 56 nucleotides. OTUs clustered via this algorithm may differ by as little as a single nucleotide and are sometimes referred to as exact sequence variants, or ESVs [62]. There is an ongoing dialog regarding the use of ESVs for fungi; for a justification of our approach see the Supplemental Material.
Chimeras were detected using the ‘uchime3 denovo’ algorithm and removed. For ITS sequences, those reads that did not merge were concatenated and processed separately, but identically, as those that did merge. OTUs made from shorter, merged reads were aligned to those made using the longer, concatenated reads and any short OTUs that aligned were not considered. We chose this approach for ITS reads because in preliminary work we discovered that, for some taxa, the ITS1 region was too lengthy to allow paired-end sequences to merge. After removing chimeras, OTUs, both long and short, were combined and an OTU table was made via aligning both merged and concatenated, unmerged reads to OTUs using the ‘usearch global’ algorithm.
Taxonomic hypotheses for OTUs were generated using the SINTAX algorithm [63] and the UNITE (v7.2; [64]) and Ribosomal Database Project database (RDP; v16; [65]) for fungi and bacteria, respectively. Read counts for OTUs corresponding to the ISD were summed for each replicate to allow normalization by the ISD. For 16S data, host plastid DNA was identified through comparison to all fabaceous chloroplasts available from the NCBI nucleotide database (accessed Feb 9, 2016; [42]). For ITS data, host DNA was identified through matching known A. lentiginosus and Oxytropis sp. (sister taxon to Astragalus) ITS sequences to OTUs. For both 16S and ITS datasets, reads for all plant OTUs were summed. In addition, for the 16S data, OTUs for mitochondrial DNA were identified using the MIDORI database [66] and removed from the OTU table. For both 16S and ITS data, those few OTUs that were not classified to any taxon were removed from the data. If >5% of the total reads for an OTU were in negative controls, then these OTUs were deemed possible contaminants and discarded (30 fungal OTUs and 2 bacterial OTUs were discarded). OTUs corresponding to A. fulva were identified through comparison to GenBank accession JX827264.1 and those corresponding to Leveillula taurica with accession MT472005.1.
Statistical analysis
We analyzed sequence count data via a hierarchical Bayesian modeling (HBM) framework that provides estimates of proportional relative abundance for each microbial taxon [67, 68]. The model estimates parameters of replicate-specific, multinomial distributions that describe taxon proportions (p parameters) and Dirichlet parameters that describe proportion estimates for the entire sampling group. This method shares information among replicates for more accurate parameter estimation and allows propagation of uncertainty in parameter estimates to downstream analyses (for a full description, see [68]). Rarefaction is not needed when using this modeling approach, because proportion estimates are used for downstream analyses and because estimates for each replicate are informed by data from all other replicates due to the hierarchical nature of the model. Modeling was conducted in the R computing environment (R Core Team [69]) using the CNVRG v0.2R package [70]. CNVRG is a user-friendly wrapper for implementing Dirichlet-multinomial modeling with rStan [71], which itself is an interface to the Stan model specification software [72]. We used the Hamiltonian Monte Carlo sampling algorithm to characterize posterior probability distributions (PPDs). We took 1000 samples from posteriors, with a thinning rate of two, after a burn-in of 500 samples. Model convergence was confirmed via the Gelman-Rubin statistic (all parameters had a statistic very near one [73]). To account for compositionality, we divided the proportion estimate for each taxon in a replicate by the proportion of reads assigned to the ISD for that replicate. By placing the relative abundances of all taxa on the scale of the ISD, we were able to better compare taxon abundances among treatment groups (for more details of the problem of compositionality and how an ISD can help see [57, 74]).
To measure the extent to which OTUs differed in relative abundance among treatment groups, PPDs for Dirichlet parameters for each OTU and treatment group were subtracted. This generated a PPD of the difference in that parameter between any two treatment groups. If 5% or less of the density of that PPD was on either side of zero, then we deemed a treatment-associated shift in microbial relative abundance was credible. Means of PPDs for parameters of interest were used as point estimates for those parameters.
Species equivalents of Shannon’s entropy and Simpson’s diversity [75] were calculated using CNVRG for the treatment group as a whole and for each replicate. To estimate diversity equivalencies for a treatment group, the equivalency was calculated for each sample of the Dirichlet distribution characterizing microbial relative abundances within that treatment group. This generated a PPD of diversity, thus propagating uncertainty in relative abundance estimates into estimates of treatment group diversity (for a similar approach see [76]). To determine how diversity equivalents differed between treatment groups, the overlap of PPDs for each group was examined (as per above). Diversity equivalents were also estimated for each replicate so that these estimates could function as the response in a linear model testing for associations between plant trait variation and shifts in microbial diversity (see below). To estimate diversity for each replicate, the means of PPDs of multinomial parameters for that replicate were calculated (recall that these parameters estimated proportional microbial relative abundance) and diversity equivalencies were calculated for the resulting vector.
HBM was also used to estimate differences among treatment groups in the mean values of plant traits, sequence-based estimates of microbial diversity, and culture richness. Each response variable was modeled as a draw from a normal distribution characteristic of the sampling group as per [77]. The mean (μ) and variance (σ2) of this distribution was estimated through sharing of information among replicates. The prior distribution for μ was a normal distribution centered at zero with a precision of 0.0001 (variance = 10,000). The prior distribution for σ2 was a uniform distribution from 0 to 100 (for full model specification see provided R code). MCMC sampling and tests for credible effects of treatment via PPD overlap were conducted as described above. For these analyses, we used the JAGS model specification language [78] as implemented via rjags v4-6 [79].
To evaluate associations between plant traits and microbial diversity, linear models were created in a HBM framework. Beta coefficients for plant traits were estimated for each treatment group, with a prior sampled from a normal distribution centered at the estimated across-treatment effect of each trait and a precision estimated across all treatments. Hyperpriors for beta coefficients were normal distributions centered at zero with a precision of 0.0001 (for full model specification see R code provided; also see [42]). Means of PPDs for each beta coefficient were used as point estimates of the effect of that covariate. The proportion of the PPD for each beta coefficient that did not overlap zero was used to determine certainty of a non-zero effect. Prior to modeling, missing values in covariates were imputed using the random-forest algorithm [80] as implemented by the randomForest R package [81]. When models were run without imputing data, results were similar to those reported here. To determine effects of treatment on microbial assemblages, as a whole, principal coordinates analysis (PCoA) and PERMANOVA were conducted on Bray–Curtis transformed tables of point estimates of proportions (derived as described above) for microbial taxa.
We chose not to report effects of treatment on endophyte richness using sequence data. To explain, when a dominant taxon is present (such as A. fulva or L. taurica) within a replicate it captures much of the sequencer’s bandwidth for that replicate. Therefore, that replicate would have fewer reads available to allocate to the other taxa present, which would result in spuriously lower richness. Consequently, to assay effects of treatment on richness, we relied on culturing data.
We omitted from all analyses ten plants for which seed coat removal did not reduce A. fulva as ascertained via swainsonine concentration (this compound is not known to be produced by the host plant), culturing, or molecular data. We also omitted plants with no evidence of A. fulva occurrence from the A. fulva positive treatment group, because A. fulva is known to be incompletely transmitted between generations [82]. Analyses were repeated while retaining all of these plants in their original treatment groups and the results obtained were qualitatively similar to those presented here.
Results
Sequencing summary and microbial diversity description
After removing host, ISD, coligo (oligos for accounting of cross-contamination), and contaminant reads and applying our stringent quality control approach, we retained for analysis 7,417,832 reads from 2292 fungal OTUs and 76,900 reads from 642 bacterial OTUs (from over 20 million 16S reads, most of which were from host organelles; for full details see the Supplemental Material).
The majority of fungal OTUs (which were specified at single nucleotide variation resolution) belonged to the Ascomycota (86%). Many OTUs were assigned to L. taurica (991 OTUs) and A. fulva (101 OTUs). A variety of bacterial taxa were observed, including many members of Acidobacteria, Actinobacteria, Proteobacteria, Firmicutes, and Chloroflexi. The most abundant bacterial OTUs were assigned to the Lactobacillaceae, Bacillaceae, Listeriaceae, and Staphylococcaceae.
Effects of the vertically transmitted fungus on the host and co-occurring microbes
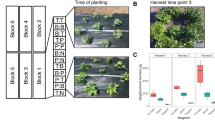
Treatment to reduce the relative abundance of the dominant, heritable fungus A. fulva from A. lentiginosus plants was successful as evidenced by read counts (Table S1), swainsonine concentrations (Fig. 1), and culturing (Fig. 2). A. fulva presence influenced plant phenotype–colonized plants were much smaller and had fewer leaves than uncolonized plants (Fig. 1, S1). The negative effect of A. fulva on plant size was observed in the second year of monitoring as well (Fig. S2). Foliar N was affected by both A. fulva and rare microbes–plants without A. fulva and that were untreated with inoculum had elevated %N in their leaves. Moreover, A. fulva generally increased the δ15 (ratio of N isotopes) and reduced NDFA, a proxy for rhizosphere nitrogen fixation activity, though these effects were less pronounced than some other effects on host phenotype (Table 2). The effects of A. fulva colonization were generally similar for plants grown from embryos planted alongside A. fulva infected agar (see Methods, for details of this control treatment), thus the observed results are not due to the confounding influence of seed coat removal (Fig. S1). We did not observe an effect of A. fulva colonization on %C, or phenology (using Fisher’s exact test to examine flowering status at time of harvest).
a + and − symbols on the x axis represent treatment to reduce the relative abundance of the vertically transmitted fungus, Alternaria fulva. Boxes shaded blue denote treatment with endophyte inoculum slurry and boxes shaded yellow denote plants that did not receive the slurry. Percentage of N, C, and swainsonine refer to foliar dry mass composition. Differences in mean trait values among treatment groups were determined through a hierarchical Bayesian analysis. Credible differences (≥95% difference in posterior probability distributions of parameter estimates) among treatment groups are designated through the letters above each boxplot. For estimates of mean trait values for each treatment group see Table 2. Boxplots summarize the data and describe interquartile range with a horizontal line denoting the median. Whiskers extend to the 10th and 90th percentiles. b shows an overview of the experimental installation and c depicts the inflorescence of A. lentiginosus. All photos by J. Harrison (color figure online).
Boxes shaded blue denote treatment with endophyte inoculum slurry; boxes shaded yellow denote plants that did not receive the slurry. The top row contains data from culturing microbes from leaves of host plants. Data shown include culture richness (a), the proportion of leaves infected with microbes other than A. fulva (b), and the rate of A. fulva infection (c). Cultured taxa were not identified to species, but were nearly all fungi. The middle row depicts the effect of treatment on Alternaria fulva (d) and Leveillula taurica (e) abundance (estimated as the ratio of the proportion of reads for either taxon to the internal standard [ISD]); these were the two most abundant fungal taxa present. The bottom row shows abundances of fungal (f) and bacterial (g) taxa that were in the inoculum mixture used to treat plants. Credible differences among treatment groups are denoted through the letters above each boxplot. Boxplots summarize the data and describe interquartile range with a horizontal line denoting the median. Whiskers extend to the 10th and 90th percentiles. Treatment to reduce Alternaria fulva is shown as A. fulva –, with a + denoting lack of treatment to reduce this taxon (color figure online).
A. fulva presence modestly affected diversity of co-occurring bacterial and fungal endophytes (Fig. 3 and Table S2). Species equivalents of Shannon’s entropy for both bacteria and fungi increased in plants colonized by A. fulva (Fig. 3), but the opposite was true for equivalents of Simpson’s diversity. Simpson’s diversity index places more weight on abundant taxa than does the Shannon index [83].
Effect of treatment on fungal (a) and bacterial diversity (b) as estimated from sequencing data. Treatments are noted on the x axis. Points denote the means of posterior probability distributions (PPD) of diversity entropy equivalencies. 95% high density intervals for these PPDs were very narrow but are shown superimposed on each estimate. Points shaded blue denote treatment groups receiving the endophyte inoculum slurry; points shaded yellow denote groups that did not receive the slurry. Treatment to reduce Alternaria fulva is shown as A. fulva –, with a + denoting lack of treatment to reduce this taxon (color figure online).
We also observed a negative association between A. fulva and L. taurica. L. taurica is a powdery mildew (Erysiphaceae) known to colonize numerous plant species, including A. lentiginosus [42]. L. taurica was the most abundant fungus sequenced and dropped in relative abundance when A. fulva was present (Fig. 2 and Table S1). This negative association was also observed visually, as we noted a powdery mildew infection on the leaves of a subset of the plants used for this experiment, and infections were less severe in plants colonized by A. fulva (Fig. S3).
For less abundant fungi (those less than 0.04% of total reads), which accounted for all fungal OTUs aside from A. fulva and L. taurica, we observed a modest increase in absolute abundance when A. fulva was present (Fig. 4), which was likely responsible for the increase in Shannon’s diversity with A. fulva infection.
The top row shows the ratio of bacterial (a) and fungal (b) proportions to the internal standard (ISD; not including A. fulva and L. taurica, which were more abundant), with proportions for all taxa summed by treatment group. The proportion of reads for all bacterial taxa was less than 3% for each treatment group (the other reads were from host organelle DNA). As for fungi, the most abundant taxon, besides A. fulva and L. taurica, was represented by less than 5% of the fungal reads in any given treatment group and, when averaged across groups, the relative abundance of this taxon was less than a percent. Most fungal taxa in most treatment groups composed less than 0.02% of reads. The average abundances of fungal phyla by treatment group are shown on the middle row (c) and abundances of bacterial phyla are shown on the bottom row (d). Blue shading denotes treatment groups receiving the endophyte inoculum slurry, whereas groups that did not receive the slurry are shaded yellow. Boxplots summarize the data and describe interquartile range with a horizontal line denoting the median. Whiskers extend to the 10th and 90th percentiles. Treatment to reduce Alternaria fulva is shown as A. fulva –, with a + denoting lack of treatment to reduce this taxon (color figure online).
The influence of horizontally transmitted endophytes on hosts
The inoculum slurry applied to plants was created from 20 morphologically distinct cultures derived from shrubs common in the Great Basin Desert. Sequencing revealed the slurry was composed of 20 fungal and 260 bacterial OTUs, including many taxa that have been observed in wild-collected plants (for a full description, see the Supplemental Material). Inoculation was successful, as shown by the effects of treatment discussed elsewhere. Additional evidence for inoculation success was that 55 of the bacterial OTUs and 3 of the fungal OTUs were sequenced from plants used in this experiment and in almost all cases, inoculum application led to an increase in the read counts obtained for those taxa (in 87% of bacterial taxa and two out of the three fungal taxa). Fungi present in inoculum that were also observed in treated plants included Preussia sp. (the most abundant fungus in the inoculum), L. taurica, and Penicillium sp., while successful bacterial colonizers were predominantly members of Proteobacteria, Actinobacteria, and Firmicutes. Inoculum application had modest effects on overall Shannon’s and Simpson’s diversity (Fig. 3). Inoculum application did not lead to significantly different centroids in PCoA ordinations (Figs. S4 and S5).
Inoculum application had no visibly pathogenic effects on plants—they appeared healthy and leaves had no evidence of necrosis. However, inoculum application did influence plant phenotype, but only when the dominant fungus A. fulva was not present. For instance, inoculum application reduced leaf count (by approximately 50%) and foliar %N (Table 2), but this was only apparent for plants without A. fulva (Fig. 1). Inoculum treatment had minor, idiosyncratic effects on trait variation, sometimes reducing and other times increasing variation (Table S3). For %N and plant size, the directionality of the effect of inoculum on trait variation depended on A. fulva treatment, with inoculation increasing trait variation when A. fulva was reduced, but decreasing it otherwise. In general, associations between the diversity of horizontally transmitted endophytes, either bacterial or fungal, with plant trait variation were weak and often limited to a specific treatment group (Tables S4 and S5).
Discussion
Foliar endophyte assemblages are typically composed of a few dominant taxa and numerous taxa of low relative abundances that occupy a small proportion of their host’s tissues (we refer to these as rare taxa [13, 84]). Ecological relevance is often considered the domain of abundant microbial taxa, because of their greater biomass and prevalence. However, our results suggest that characterizing the overall effects of endophyte assemblages may require study of rare taxa. Indeed, we report that a suite of rare endophytes affected host size and foliar N content, among other traits. We also observed that the influence of these taxa was attenuated by the presence of a dominant fungal endophyte, which itself mediated host plant phenotype.
It is important to note that, regardless of treatment, host plants appeared healthy to the eye—their tissues were green, and, in many cases, they fruited successfully during the first and second years of growth. Thus, both A. fulva and co-occurring microbes meet the criterion of living asymptomatically within plant tissues necessary for designation as endophytic taxa rather than obligate pathogens [3].
Aside from A. fulva and L. taurica, all other microbial taxa observed, be they fungal or bacterial, were rare, as shown through read counts (Fig. 4) and culture-determined infection rate (Fig. 2b). Microbial taxa within the inoculum mixture were of similarly very low abundance in plants (Fig. 2). Indeed, the abundance of all these taxa combined was generally about that of the ISD, which was spiked into samples with the minuscule concentration of 0.03 pg/μL. Thus, it was surprising that we saw fairly dramatic effects of treatment with the inoculum.
We were not able to attribute the effects of the inoculum to specific taxa, but this was by design. We were curious what the overall effects of a complex endophyte assemblage might be for plants grown in nature—thus, we were attempting to provide a different perspective than that offered by gnotobiotic studies in laboratory settings. Many of the taxa in our inoculum mixture have been observed in wild plants, with low relative abundances [42], including possible pathogens such as Preussia. Thus, it seems very plausible that the effects of the inoculum mixture that we observed could occur in wild populations of A. lentiginosus.
It seems unlikely that unintended consequences of treatment induced the results we observed since we applied a mock treatment to account for the effects of leaf wetting and surfactant application and we controlled for the effect of embryo excision from seed coats. However, we considered two caveats to our results. First, it is possible that our inoculum mixture was biased toward endophytes that grow rapidly in culture. If that was the case, then perhaps an inoculum mixture containing slower-growing taxa, which plausibly could include more biotrophic taxa, would have different effects on the host than those we observed. Second, our plants were reared with regular watering. This was by necessity because the plants were grown in pots as opposed to planted directly in the soil. Pots provided many logistical advantages, but their use led to a heating of the soil and a rapid draining of water that would have killed many plants, if they did not receive water that was supplemental to precipitation. It is unclear what effects watering might have had on our results, though we note that we were careful to avoid wetting leaves during watering as leaf wetness can influence microbial colonization.
Ecological roles of rare endophytes
The effects of rare endophytes that we observed likely have implications for host fitness. Certainly the approximately 50% reduction in plant size and leaf number that we report could lead to reduced seed output (Table 2) and it seems likely that the effects on foliar N and leaf morphology we observed could also influence host fitness. Beyond the direct influence of trait variation to plant persistence and reproductive output, the marked shifts in phenotype that we observed could have indirect effects on host-associated organisms, such as arthropods, which also affect host fitness. For instance, size and foliar %N are often strong predictors of variation in insect assemblages and herbivory across plant species [85, 86], thus shifts in these traits induced by low-biomass microbial taxa could have cascading effects on arthropod communities.
We considered two possibilities for the distribution of ecological influence among rare endophytic taxa. Specifically, influence could be limited to several keystone taxa or could be cumulative, such that a quorom must be reached before the combined effect of rare endophytes induces a response by the host. We were unable to satisfactorily resolve these two non-mutually exclusive hypotheses. However, the quorom hypothesis might lead to a negative association between microbial diversity and plant size, because higher microbial diversity would occur when taxa were more evenly distributed, each with a role to play. We did not unequivocally observe such a negative association (Tables S4 and S5). Indeed, we found that most rare endophytes occurred infrequently in samples, suggesting that if a quorom was present and responsible for the shifts in host phenotype observed, then that quorom must be easily met and be composed of very little total biomass that does not manifest in notable shifts in diversity.
Alternatively, infrequently observed, keystone taxa could have caused the treatment effects we report, as these taxa, by definition, exert greater influence than would be predicted from their low biomass. For instance, a localized infection by a keystone taxon could have effects that spread throughout the host (e.g., through hormone stimulation), yet that taxon would not be present in the majority of leaves sequenced from that host. This concept suggests limitations of the common practice of in silico identification of keystone taxa as those taxa that are prevalent among samples, such that their removal from co-occurrence networks causes a shift in network topology [87,88,89]. We reiterate the non-exclusivity of the keystone and quorom hypotheses and suggest disentangling the two represents a profitable line of inquiry for future work.
Effect of Alternaria fulva, the heritable fungus
In addition to the influence of rare endophytes, we observed that the dominant, vertically transmitted fungal endophyte A. fulva also reduced host size and foliar %N (Fig. 1; consistent with Cook et al. [51]). Inhibition of host growth by A. fulva is perplexing, because the fungus is vertically transmitted in seeds and, therefore, its fitness is tied to that of its host. Larger A. lentiginosus plants generally produce more seeds (J. Harrison, personal observation) and, thus, selection should operate against mechanisms by which A. fulva reduces host growth. On the other hand, A. fulva grows very slowly in culture [90, 91] and fast-growing plants could possibly outpace hyphal growth. If the fungus cannot grow fast enough to reach seeds before they mature, then its direct fitness is zero. Consequently, constraining host growth may improve fungal fitness, because it would allow time for hyphae to reach reproductive structures. This hypothesis awaits further testing.
Another intriguing possibility is that a fungal-induced reduction in plant size could actually improve plant longevity in extreme conditions and thereby lead to a positive, time-averaged, effect on fungal fitness. For several native plants in the Great Basin, small stature paradoxically facilities the ability to withstand drought and competition from invasive annual grasses [92, 93]. Thus, it is possible that plants colonized by Alternaria endophytes could better survive the harsh desert climate, providing both the plant and the fungus more opportunity to reproduce. Interestingly, previous work has shown that Alternaria endophytes do not reduce plant size in locoweeds that are drought-stressed [94] and that swainsonine concentration can increase during drought stress. Thus, perhaps the negative affect of A. fulva on host size we observed here, in a well-watered, controlled setting, would not play out in drought-stressed wild populations.
The potential fitness costs imposed by A. fulva on its host may be ameliorated by the negative association we observed between A. fulva and the most abundant pathogen present, L. taurica (Fig. 2). These results support the hypothesis posed by Lu et al. [49] that the Alternaria spp. occurring within Astragalus and Oxytropis act as mutualists to their hosts by restricting pathogen exposure. A. lentiginosus is a plant of frequently disturbed, climatically variable, arid landscapes and it is likely that pathogen pressure in such locales is particularly damaging, because the lack of resources could impede recovery from tissue loss. The same rationale has inspired the growth-rate hypothesis in the literature characterizing interactions between plants and insect herbivores [95]. This hypothesis predicts plants growing in resource poor conditions will recuperate from herbivory slowly, and thus benefit from investment in phytochemical defenses that would otherwise be too costly. Similarly, tolls imposed by A. fulva on A. lentiginosus may be acceptable to the host given the harshness of the arid American West.
Our results compliment recent research presented in Christian et al. [30] showing that endophytes, and interactions between endophytes and pathogens, can alter N distribution and uptake in plants. The study did not demonstrate endophyte-induced shifts in %N content at the whole plant level (such as those we observed here), but it did show that endophytes influenced N uptake in plants and affected N distribution among leaves (also see [32]). We also observed credible, treatment-induced shifts in δ15N and nitrogen fixation in the rhizosphere (NDFA) (Fig. 1 and Table 2), but we also found that rare endophytes reduced total foliar %N when A. fulva was not present to attenuate their effects. When taken together with previous work [30, 32], our results suggest that the effects of horizontally transmitted endophytes on foliar N depend on host taxon and individual, abiotic context (e.g. N availability), and interactions with other microbiota, and, when these factors align, the effect of endophytes on N allocation within hosts can be noteworthy. Notably, we saw some subtle trends that bear further examination; specifically, reducing A. fulva colonization was associated with higher NDFA. Thus, the work we present here, when coupled with the results in Christian et al. [30], suggest that foliar endophytes may affect N fixation, which typically happens below ground. This suggests that interactions among microbes in the phyllosphere can influence what happens in the rhizosphere.
Other considerations: a milieu of interactions and experimental design
Our study demonstrates the ecological consequences of interactions among microbes [18], as evidenced by a negligible effect of inoculum application for plants colonized by A. fulva and the negative association between A. fulva and L. taurica. We suggest that these results are not likely due to direct competition for resources between A. fulva and other microbes–the disparity in leaf size and microbe size is too great and there seems to be enough healthy leaf tissue for all parties (based on the lack of visual infection symptoms in our plants). Even for A. fulva, which grows systemically through its host, physical encounters with co-occurring microbes are probably rare—with the possible exception of encounters with L. taurica. We note that it is plausible that swainsonine could inhibit growth of competing microbes, as the molecule inhibits the action of alpha-mannosidase [40], which is used by fungi to process oligosaccharides [96]. However, it is unknown if swainsonine is exuded into plant tissues or is instead retained within fungal cells, if the latter, then it is not clear how the compound would affect co-occurring microbes. We suggest that indirect mediation of microbe–microbe interactions by the host is more likely. Indeed, gene expression studies in several perennial grasses [33, 34, 97] and in Theobroma cacao [32] have demonstrated an upregulation in the host immune response after colonization by endophytes (see [98]). To speculate, it is possible that A. fulva similarly primes the host immune response, which could negatively affect co-occurring microbes.
To account for any adverse effects of seed coat removal, we planted control seeds alongside sterile agar or agar inoculated with A. fulva. This technique was successful as shown by culturing results (Fig. 2), swainsonine concentrations (Fig. S1), and sequencing output (Fig. 2). The results we observed from control plants were very similar to those from treated plants, except for foliar C and N concentrations, which were more variable among controls. Most manipulative studies of vertically transmitted fungal endophytes attempt to kill fungi within seeds through either heat treatment (e.g. [99]), long-term storage (e.g. [7]), or fungicide application (e.g. [100]). While studies manipulating endophytes via these techniques have been of critical importance, it is possible that these treatments could have undesirable consequences that are hard to control for and that could obscure effects of endophyte reduction. Consequently, we suggest others consider the approach we used here when seeds from endophyte-free plants are not available.
Conclusion
Our results suggest that rare, low-biomass endophytic taxa can have marked influence on their hosts and that these effects may be mediated by co-occurring dominant microbial taxa. It remains to be seen how often rare endophytic taxa affect host phenotypes in other systems. However, given that every study of endophytic biodiversity with which we are familiar shows a steep rank-abundance curve, with many taxa of low relative abundances, it seems plausible that the cumulative role of rare microbes across hosts could be substantial.
We also hope that a more careful consideration of different taxa (common and rare) within endophyte assemblages will illuminate parallels with diversity-ecosystem function studies of macroscopic organisms where community-wide effects of rare taxa have been demonstrated [101]. As biodiversity declines, such connections across scales of observation could provide impetus for conserving rare taxa, large and small, as important contributors to ecosystem processes.
Data availability
All scripts, plant trait data, and processed sequence data are available at: https://github.com/JHarrisonEcoEvo/RareMicrobes. Raw data are hosted by the University of Wyoming. MiSeq data used during preliminary work for this experiment can be found at: https://doi.org/10.15786/r9xy-6x03 while NovaSeq data analyzed herein can be found at: https://hdl.handle.net/20.500.11919/7166.
References
Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett. 2008;278:1–9. https://doi.org/10.1111/j.1574-6968.2007.00918.x.
Rodriguez R, White J Jr, Arnold A, Redman R. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182:314–30.
Wilson D. Endophyte: the evolution of a term, and clarification of its use and definition Oikos. 1995;73:274–6.
Harrison JG, Griffin EA. The diversity and distribution of endophytes across biomes, plant phylogeny and host tissues: How far have we come and where do we go from here?. Environ Microbiol. 2020;22:2107–23. https://doi.org/10.1111/1462-2920.14968.
Clay K, Schardl C. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat. 2002;160:S99–S127. https://doi.org/10.1086/342161.
Rudgers JA, Afkhami ME, Rúa MA, Davitt AJ, Hammer S, Huguet VM. A fungus among us: broad patterns of endophyte distribution in the grasses. Ecology. 2009;90:1531–9. https://doi.org/10.1890/08-0116.1.
Clay K, Holah J. Fungal endophyte symbiosis and plant diversity in successional fields. Science. 1999;285:1742–4. https://doi.org/10.1126/science.285.5434.1742.
Afkhami ME, Strauss SY. Native fungal endophytes suppress an exotic dominant and increase plant diversity over small and large spatial scales. Ecology. 2016;97:1159–69. https://doi.org/10.1890/15-1166.1.
Rudgers JA, Clay K. An invasive plant–fungal mutualism reduces arthropod diversity. Ecol Lett. 2008;11:831–40. https://doi.org/10.1111/j.1461-0248.2008.01201.x.
Gorischek AM, Afkhami ME, Seifert EK, Rudgers JA. Fungal symbionts as manipulators of plant reproductive biology. Am Nat. 2013;181:562–70. https://doi.org/10.1086/669606.
Malloch D, Blackwell M. Dispersal of fungal diaspores. The fungal community: Its organization and role in the ecosystem. 2nd ed. New York, NY: Marcel Dekker, Inc; 1992. p. 147–71.
Devarajan P, Suryanarayanan T. Evidence for the role of phytophagous insects in dispersal of non-grass fungal endophytes. Fungal Divers. 2006;23:111–9.
Lodge DJ, Fisher P, Sutton B. Endophytic fungi of Manilkara bidentata leaves in Puerto Rico. Mycologia. 1996;88:733–8.
Paine RT. A note on trophic complexity and community stability. Am Nat. 1969;103:91–93. https://doi.org/10.1086/282586.
Jenkins SH, Busher PE. Castor canadensis, Mammalian Species. 1979. https://doi.org/10.2307/3503787.
Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–25. https://doi.org/10.1038/nrmicro2873.
Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V, et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017;11:853–62. https://doi.org/10.1038/ismej.2016.174.
Hassani MA, Durán P, Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:58. https://doi.org/10.1186/s40168-018-0445-0.
Rockman MV. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution. 2012;66:1–17. https://doi.org/10.1111/j.1558-5646.2011.01486.x.
Beckers GJ, Conrath U. Priming for stress resistance: from the lab to the field. Curr Opin Plant Biol. 2007;10:425–31. https://doi.org/10.1016/j.pbi.2007.06.002.
Hartmann A, Rothballer M, Hense BA, Schröder P. Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Front Plant Sci. 2014;5. https://doi.org/10.3389/fpls.2014.00131.
Friesen ML, Porter SS, Stark SC, von Wettberg EJ, Sachs JL, Martinez-Romero E. Microbially mediated plant functional traits. Annu Rev Ecol Evol Syst. 2011;42:23–46. https://doi.org/10.1146/annurev-ecolsys-102710-145039.
Hardoim PR, Van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. https://doi.org/10.1128/MMBR.00050-14.
Doty SL. Growth-promoting endophytic fungi of forest trees. In: Pirttilä AM, Frank AC, editors. Endophytes of forest trees: biology and applications. Dordrecht: Springer Netherlands; 2011;151–6.
Arnold AE, Herre EA. Canopy cover and leaf age affect colonization by tropical fungal endophytes: Ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia. 2003;95:388–98. http://www.mycologia.org.unr.idm.oclc.org/content/95/3/388. Accessed 12 Dec 2016.
Busby PE, Peay KG, Newcombe G. Common foliar fungi of Populus trichocarpa modify melampsora rust disease severity. New Phytol. 2016;209:1681–92. https://doi.org/10.1111/nph.13742.
Christian N, Herre EA, Mejia LC, Clay K. Exposure to the leaf litter microbiome of healthy adults protects seedlings from pathogen damage. Proc R Soc B. 2017;284:20170641. https://doi.org/10.1098/rspb.2017.0641.
Cheplick GP, Cho R. Interactive effects of fungal endophyte infection and host genotype on growth and storage in Lolium perenne. New Phytol. 2003;158:183–91. https://doi.org/10.1046/j.1469-8137.2003.00723.x.
Zahn G, Amend AS. Foliar fungi alter reproductive timing and allocation in arabidopsis under normal and water-stressed conditions. 2019. https://www.biorxiv.org/content/10.1101/519678v1.
Christian N, Herre EA, Clay K. Foliar endophytic fungi alter patterns of nitrogen uptake and distribution in Theobroma cacao. New Phytol. 2019;222:1573–83. https://doi.org/10.1111/nph.15693.
Rosado BHP, Almeida LC, Alves LF, Lambais MR, Oliveira RS. The importance of phyllosphere on plant functional ecology: a phyllo trait manifesto. New Phytol. 2018;219:1145–9. https://doi.org/10.1111/nph.15235.
Mejía LC, Herre EA, Sparks JP, Winter K, García MN, Van Bael SA, et al. Pervasive effects of a dominant foliar endophytic fungus on host genetic and phenotypic expression in a tropical tree. Front Microbiol. 2014;5:479.
Dupont PY, Eaton CJ, Wargent JJ, Fechtner S, Solomon P, Schmid J, et al. Fungal endophyte infection of ryegrass reprograms host metabolism and alters development. New Phytol. 2015;208:1227–40. https://doi.org/10.1111/nph.13614.
Dinkins RD, Nagabhyru P, Graham MA, Boykin D, Schardl CL. Transcriptome response of Lolium arundinaceum to its fungal endophyte Epichloë coenophiala. New Phytol. 2017;213:324–37. https://doi.org/10.1111/nph.14103.
Welsh S, North American species of astragalus Linnaeus (Leguminosae): a taxonomic revision. Provo, Utah: Brigham Young University; 2007.
Knaus BJ. Morphometric architecture of the most taxon-rich species in the U.S. Flora: Astragalus lentiginosus (Fabaceae). Am J Bot. 2010;97;1816–26. https://doi.org/10.3732/ajb.0900145.
Baucom DL, Romero M, Belfon R, Creamer R. Two new species of undifilum, fungal endophytes of astragalus (locoweeds) in the United States. Botany. 2012;90:866–75. https://doi.org/10.1139/b2012-056.
Woudenberg JHC, Groenewald JZ, Binder M, Crous PW. Alternaria redefined. Stud Mycol. 2013;75:171–212. https://doi.org/10.3114/sim0015.
Cook D, Gardner DR, Martinez A, Robles CA, Pfister JA. Screening for swainsonine among South American astragalus species. Toxicon. 2017;139:54–7. https://doi.org/10.1016/j.toxicon.2017.09.014.
Molyneux RJ, James LF. Loco intoxication: indolizidine alkaloids of spotted locoweed (Astragalus lentiginosus). Science. 1982;216:190–1. https://doi.org/10.1126/science.6801763.
Cook D, Gardner DR, Ralphs MH, Pfister JA, Welch KD, Green BT. Swainsoninine concentrations and endophyte amounts of Undifilum oxytropis in different plant parts of Oxytropis sericea. J Chem Ecol. 2009;35:1272–8. https://doi.org/10.1007/s10886-009-9710-9.
Harrison JG, Parchman TL, Cook D, Gardner DR, Forister ML. A heritable symbiont and host-associated factors shape fungal endophyte communities across spatial scales. J Ecol. 2018;106:2274–86. https://doi.org/10.1111/1365-2745.12967.
Grum DS, Cook D, Baucom D, Mott IW, Gardner DR, Creamer R, et al. Production of the alkaloid swainsonine by a fungal endophyte in the host Swainsona canescens. J Nat Prod. 2013;76:1984–8. https://doi.org/10.1021/np400274n.
Cook D, Gardner DR, Pfister JA. Swainsonine-containing plants and their relationship to endophytic fungi. J Agric Food Chem. 2014;62:7326–34. https://doi.org/10.1021/jf501674r.
Panaccione DG, Beaulieu WT, Cook D. Bioactive alkaloids in vertically transmitted fungal endophytes. Funct Ecol. 2014;28:299–314. https://doi.org/10.1111/1365-2435.12076.
Thompson DC, Knight JL, Sterling TM, Murray LW. Preference for specific varieties of woolly locoweed by a specialist weevil, Cleonidius trivittatus (Say). Southwest Entomol. 1995;20:325–325.
Parker JE. Effects of insect herbivory by the four-lined locoweed weevil, Cleonidius trivittatus (say) (Coleoptera: Curculionidae), on the alkaloid swainsonine in locoweeds Astragalus mollissimus and Oxytropis sericea. Ph.D. thesis. Las Cruces, New Mexico: New Mexico State University; 2008.
Creamer R, Baucom D. Fungal endophytes of locoweeds: a commensal relationship? J Plant Physiol Pathol. 2013;1. https://doi.org/10.4172/2329-955X.1000104.
Lu H, Quan H, Zhou Q, Ren Z, Xue R, Zhao B, et al. Endogenous fungi isolated from three locoweed species from rangeland in western China. Afr J Microbiol Res. 2017;11:155–70. https://doi.org/10.5897/AJMR2016.8392.
Schulthess FM, Faeth SH. Distribution, abundances, and associations of the endophytic fungal community of Arizona fescue (Festuca arizonica). Mycologia. 1998;90:569–78. https://doi.org/10.1080/00275514.1998.12026945.
Cook D, Gardner DR, Pfister JA, Stonecipher CA, Robins JG, Morgan JA. Effects of elevated CO2 on the swainsonine chemotypes of Astragalus lentiginosus and Astragalus mollissimus. J Chem Ecol. 2017;43:307–16. https://doi.org/10.1007/s10886-017-0820-5.
Oldrup E, McLain-Romero J, Padilla A, Moya A, Gardner D, Creamer R. Localization of endophytic undifilum fungi in locoweed seed and influence of environmental parameters on a locoweed in vitro culture system. Botany. 2010;88:512–21. https://doi.org/10.1139/B10-026.
Gardner DR, Molyneux RJ, Ralphs MH. Analysis of swainsonine: extraction methods, detection, and measurement in populations of locoweeds (oxytropis spp.). J Agric Food Chem. 2001;49:4573–80.
Högberg P. 15N natural abundance in soil–plant systems. New Phytol. 1997;137:179–203. https://www.cambridge.org/core/journals/new-phytologist/article/tansley-review-no-95-15n-natural-abundance-in-soilplant-systems/304069FD5C8283EDB78D0AA594465E71. Accessed 2 Jul 2017.
Wang Y, Qian P-Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE. 2009;4:e7401. https://doi.org/10.1371/journal.pone.0007401.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In Innis MA, Glefand DH, Sninsky JJ, White TJ, editors, PCR protocols: a guide to methods and applications. London: Academic Press; 1990.
Harrison JG, Calder WJ, Shuman B, Buerkle CA, The quest for absolute abundance: the use of internal standards for DNA-based community ecology. Mol Ecol Resour. 2020, https://doi.org/10.1111/1755-0998.13247.
Tourlousse DM, Yoshiike S, Ohashi A, Matsukura S, Noda N, Sekiguchi Y. Synthetic spike-in standards for high-throughput 16S rRNA gene amplicon sequencing. Nucleic Acids Res. 2017;45:e23–e23. https://doi.org/10.1093/nar/gkw984.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
Rognes T, Flouri T, Nichols B, Quince C, Mahé F, “VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016. https://doi.org/10.7717/peerj.2584.
Edgar RC, UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. 2016. https://www.biorxiv.org/content/10.1101/081257v1.full.
Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017:2639. https://doi.org/10.1038/ismej.2017.119.
Edgar R, “SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. 2016. https://www.biorxiv.org/content/10.1101/074161v1.
Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, https://doi.org/10.1093/nar/gky1022.
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis Nucleic Acids Res. 2014. 42, https://doi.org/10.1093/nar/gkt1244.
Machida RJ, Leray M, Ho S-L, Knowlton N. Metazoan mitochondrial gene sequence reference datasets for taxonomic assignment of environmental samples. Sci Data. 2017;4:17007 https://doi.org/10.1038/sdata.2017.27.
Fordyce JA, Gompert Z, Forister ML, Nice CC. A hierarchical Bayesian approach to ecological count data: A flexible tool for ecologists. PLoS ONE. 2011;6;e26785. https://doi.org/10.1371/journal.pone.0026785.
Harrison JG, Calder WJ, Shastry V, Buerkle CA. Dirichlet-multinomial modelling outperforms alternatives for analysis of microbiome and other ecological count data. Mol Ecol Resour. 2020;20:481–97. https://doi.org/10.1111/1755-0998.13128.
R Core Team, R: a language and environment for statistical computing. Vienna: R Core Team; 2019.
Harrison J, Shastry V, Calder WJ, Buerkle CA, “CNVRG: Dirichlet-multinomial modelling of relative abundance data.” Sep. 2020. https://CRAN.R-project.org/package=CNVRG. Accessed 28 Oct 2020.
S. D. Team, Stan modeling language users guide and reference manual. 2020. https://mc-stan.org/users/documentation/
S. D. Team, “RStan: The R interface to Stan. R package.” 2020. http://mc-stan.org/.
Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;7:457–72. http://www.jstor.org/stable/2246093. Accessed 16 Jun 2018.
Gloor GB, Macklaim JM, Vu M, Fernandes AD. Compositional uncertainty should not be ignored in high-throughput sequencing data analysis. Austrian J Stat. 2016;45:73–87. http://ajs.data-analysis.at/index.php/ajs/article/view/vol45-4-5. Accessed 4 Dec 2017.
Jost L. Entropy and diversity. Oikos. 2006;113:363–75.
Marion ZH, Fordyce JA, Fitzpatrick BM. A hierarchical Bayesian model to incorporate uncertainty into methods for diversity partitioning. Ecology. 2018;99:947–56. https://doi.org/10.1002/ecy.2174.
Harrison JG, Gompert Z, Fordyce JA, Buerkle CA, Grinstead R, Jahner JP. et al. The many dimensions of diet breadth: phytochemical, genetic, behavioral, and physiological perspectives on the interaction between a native herbivore and an exotic host. PLoS ONE. 2016;11:e0147971. https://doi.org/10.1371/journal.pone.0147971.
Plummer M. JAGS: a program for analysis of Bayesian graphical models using Gibbs sampling,”. Proc 3rd Int workshop Distrib Stat Comput. 2003;124:1–8. 10.2003.
Plummer M. Rjags: Bayesian graphical models using MCMC. R package version 3-15. 2015. Https://CRAN.R-project.org/package=rjags.
Breiman L. Random forests. Mach Learn. 2001;45:5–32. https://doi.org/10.1023/A:1010933404324.
Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22.
Grum DS, Cook D, Gardner DR, Roper JM, Pfister JA, Ralphs MH. Influence of seed endophyte amounts on swainsonine concentrations in astragalus and oxytropis locoweeds. J Agric Food Chem. 2012;60:8083–9. https://doi.org/10.1021/jf3024062.
Marion ZH, Fordyce JA, Fitzpatrick BM. Extending the concept of diversity partitioning to characterize phenotypic complexity. Am Nat. 2015;186:348–61.
Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots?”. Ecology. 2007;88:541–49. https://doi.org/10.1890/05-1459.
Strong DR, Lawton JH, Southwood SR. Insects on plants. Community patterns and mechanisms. Oxford, UK: Blackwell Scientific Publicatons; 1984.
Carmona D, Lajeunesse MJ, Johnson MTJ. Plant traits that predict resistance to herbivores. Funct Ecol. 2011;25:358–67. https://doi.org/10.1111/j.1365-2435.2010.01794.x.
Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol. 2014;5. https://doi.org/10.3389/fmicb.2014.00219.
Trosvik P, de Muinck EJ. Ecology of bacteria in the human gastrointestinal tract—identification of keystone and foundation taxa. Microbiome. 2015;3:44 https://doi.org/10.1186/s40168-015-0107-4.
Banerjee S, Schlaeppi K, van der Heijden MGA, Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16:567. https://doi.org/10.1038/s41579-018-0024-1.
Braun K, Romero J, Liddell C, Creamer R. Production of swainsonine by fungal endophytes of locoweed. Mycological Res. 2003;107:980–8. https://doi.org/10.1017/S095375620300813X.
Noor AI, Nava A, Cooke P, Cook D, Creamer R. Evidence for nonpathogenic relationships of alternaria section undifilum endophytes within three host locoweed plant species. Botany. 2018;96:187–200. https://doi.org/10.1139/cjb-2017-0117.
Kulpa SM, Leger EA. Strong natural selection during plant restoration favors an unexpected suite of plant traits. Evolut Appl. 2013;6:510–23. https://doi.org/10.1111/eva.12038.
Leger EA, Baughman OW. What seeds to plant in the Great Basin? Comparing traits prioritized in native plant cultivars and releases with those that promote survival in the field. Nat Areas J. 2015;35:54–68. https://doi.org/10.3375/043.035.0108.
Klypina N, Pinch M, Schutte BJ, Maruthavanan J, Sterling TM, Water-deficit stress tolerance differs between two locoweed genera (astragalus and oxytropis) with fungal endophytes. Weed Sci. 2017:1–13. https://doi.org/10.1017/wsc.2017.21.
Stamp N. Out of the quagmire of plant defense hypotheses. Q Rev Biol. 2003;78:23–55. https://doi.org/10.1086/367580.
Eades CJ, Hintz WE. Characterization of the α-mannosidase gene family in filamentous fungi: N-glycan remodelling for the development of eukaryotic expression systems. Biotechnol Bioprocess Eng. 2000;5:227. https://doi.org/10.1007/BF02942178.
Schmid J, Day R, Zhang N, Dupont PY, Cox MP, Schardl CL, et al. Host tissue environment directs activities of an epichloë endophyte, while it induces systemic hormone and defense responses in its native perennial ryegrass host. Mol Plant Microbe Interact. 2016;30:138–49. https://doi.org/10.1094/MPMI-10-16-0215-R.
Zamioudis C, Pieterse CMJ. Modulation of host immunity by beneficial microbes. Mol Plant-Microbe Interact. 2011;25:139–50. https://doi.org/10.1094/MPMI-06-11-0179.
Kannadan S, Rudgers JA. Endophyte symbiosis benefits a rare grass under low water availability. Funct Ecol. 2008;22:706–13. https://doi.org/10.1111/j.1365-2435.2008.01395.x.
Barillas JRV, Paschke MW, Ralphs MH, Child RD. White locoweed toxicity is facilitated by a fungal endophyte and nitrogen-fixing bacteria. Ecology. 2007;88:1850–6. https://doi.org/10.1890/06-0728.1.
Isbell F, Calcagno V, Hector A, Connolly J, Harpole WS, Reich PB, et al. High plant diversity is needed to maintain ecosystem services. Nature. 2011;477:199–202. https://doi.org/10.1038/nature10282.
Acknowledgements
We received three very insightful reviews that improved this manuscript substantially. Our thanks go to these reviewers. Funding was provided by a National Science Foundation (NSF) Doctoral Dissertation Grant awarded to JGH. JGH and CAB were supported by the Microbial Ecology Collaborative with funding from NSF award #EPS-1655726, and MLF was supported with funding from NSF award #DEB-1638793. Computing was performed in the Teton Computing Environment at the Advanced Research Computing Center, University of Wyoming, Laramie (https://doi.org/10.15786/M2FY47).
Author information
Authors and Affiliations
Contributions
JGH, LPB, and MLF conducted the field experiment. LPB and JGH performed culturing. JGH executed analyses. Analytical chemistry was conducted by DC and DRG. All authors contributed to experimental design and manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Harrison, J.G., Beltran, L.P., Buerkle, C.A. et al. A suite of rare microbes interacts with a dominant, heritable, fungal endophyte to influence plant trait expression. ISME J 15, 2763–2778 (2021). https://doi.org/10.1038/s41396-021-00964-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-00964-4
This article is cited by
-
Minor species of foliar fungal endophyte communities: do they matter?
Mycological Progress (2021)