Abstract
Objective
Brain atrophy has the potential to become a biomarker for severity of radiation-induced side-effects. Particularly brain tumour patients can show great MRI signal changes over time caused by e.g. oedema, tumour progress or necrosis. The goal of this study was to investigate if such changes affect the segmentation accuracy of normal appearing brain and thus influence longitudinal volumetric measurements.
Materials and methods
T1-weighted MR images of 52 glioblastoma patients with unilateral tumours acquired before and three months after the end of radio(chemo)therapy were analysed. GM and WM volumes in the contralateral hemisphere were compared between segmenting the whole brain (full) and the contralateral hemisphere only (cl) with SPM and FSL. Relative GM and WM volumes were compared using paired t tests and correlated with the corresponding mean dose in GM and WM, respectively.
Results
Mean GM atrophy was significantly higher for full segmentation compared to cl segmentation when using SPM (mean ± std: ΔVGM,full = − 3.1% ± 3.7%, ΔVGM,cl = − 1.6% ± 2.7%; p < 0.001, d = 0.62). GM atrophy was significantly correlated with the mean GM dose with the SPM cl segmentation (r = − 0.4, p = 0.004), FSL full segmentation (r = − 0.4, p = 0.004) and FSL cl segmentation (r = -0.35, p = 0.012) but not with the SPM full segmentation (r = − 0.23, p = 0.1).
Conclusions
For accurate normal tissue volume measurements in brain tumour patients using SPM, abnormal tissue needs to be masked prior to segmentation, however, this is not necessary when using FSL.
Similar content being viewed by others
Introduction
Radiotherapy is part of the standard treatment regime of gliomas. With improving survival in particular for low-grade tumours, the number of patients experiencing therapy-induced long-term complications, such as neurocognitive decline, increases accordingly [1,2,3]. Apart from subjective measures, such as patient-reported outcomes and neurocognitive function tests, the community strives to include objective measures in the evaluation of treatment outcome, especially when comparing different treatment modalities, such as photon- and proton-based radio(chemo)therapy.
Radiotherapy induced brain atrophy has been observed across the whole brain [4,5,6,7], hippocampus [8,9,10,11], amygdala [12] and cerebellum [13]. The findings show consistently that the amount of tissue loss is dose dependent [5, 13, 14]. Previous studies have also linked atrophy with cognitive decline [4, 15]. Brain atrophy has become an important marker of disease severity in neurodegenerative and demyelinating diseases such as Alzheimer’s disease [16] or multiple sclerosis (MS) [17]. Similarly, atrophy has the potential to become a biomarker for severity of radiation-induced side effects [4, 15]. However, this requires measurement of the normal tissue volumes of brain tumour patients with great accuracy across multiple time points. Particularly brain tumours can show great changes in MR signal intensities over time in and around the original tumour site caused by oedema, tumour progression, or treatment itself. If these signal changes influence the atrophy measurement, they could mask its potential to act as a reliable biomarker.
In the research environment, accurate measurement of atrophy commonly involves segmentation of high resolution T1-weighted MR images into grey matter (GM), white matter (WM) and cerebrospinal fluid (CSF) using software such as FSL (FMRIB Software Library: https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) [18, 19] or SPM (Statistical Parametric Mapping: https://www.fil.ion.ucl.ac.uk/spm/) [20, 21]. These software tools use an iterative Bayesian segmentation approach. By default, FSL initializes the tissue segmentation using intensity-based thresholds to segment the input image coarsely into three tissue types corresponding to GM, WM and CSF [18]. SPM uses prior tissue probability maps (TPMs) to initialize the tissue segmentation. The TPMs are derived from a collection of manually segmented healthy brains [20]. Consequently, if abnormal brain tissue is present, for instance, resection cavities, oedema, and tumour tissue in brain tumour patients, or WM lesions in MS patients, the TPMs are not valid within these regions of abnormal tissue and can cause significant bias in the resulting GM and WM segmentation. Abnormal tissue will be incorrectly classified as either GM, WM, or CSF. Consequently, image intensities of abnormal tissue contribute to the global joint-probability distribution model that is constructed for each of the GM, WM, and CSF tissues during the iterative process of segmentation. This can alter the width and position of the respective GM, WM, or CSF probability density functions on the MRI intensity scale and consequently lead to a shift between the separation of tissue boundaries, most commonly between GM/WM and GM/CSF, thus biasing volume measurements in the normal tissue even when abnormal tissue is removed after segmentation.
Segmenting MR images containing abnormal tissue has been studied previously in the field of MS, where WM lesions can have a significant impact on GM and WM volumes [22,23,24]. In MS, the error in volume measurements mainly arises from misclassification of WM lesions as GM or CSF [25]. Brain tumours, conversely, generally have much larger lesions. Using Bayesian segmentation approaches, this could also potentially bias normal tissue volume measurements if the abnormal tissue is not masked prior to segmentation.
The aim of this study was to assess the influence of the presence of residual tumour, resection cavities, and oedema on the quality of the tissue segmentation in the healthy tissue and on the longitudinal assessment of volumetric changes in brain parenchyma. To that end, we measured GM and WM volumes segmented from 3D 1 mm isotropic T1-weighted MR images in a cohort of glioblastoma patients with unilateral lesions. We assessed the longitudinal volumetric changes between the baseline and 3 month follow-up after radio(chemo-)therapy, respectively. We hypothesized that the presence of abnormal tissue in the ipsilateral hemisphere would affect the segmentation and longitudinal volumetric changes in the healthy contralateral hemisphere. To show that, we compared segmentations obtained from SPM and FSL with and without removing the tumour-bearing hemisphere.
Materials and methods
Subjects
Patient data were acquired as part of a prospective longitudinal study investigating the effect of 11C-methionine PET/MR for tailoring the treatment of patients with glioblastoma, approved by the local ethics committee (NCT01873469, EK41022013, BO-EK-167052020). Gross tumour resection was performed in most patients prior to radio(chemo-)therapy. Baseline MR images were acquired two to seven weeks after surgery and typically two weeks before the start of radio(chemo-)therapy. Radiotherapy was conducted using either photon or proton therapy as described previously [26]. Follow-up MRIs were acquired approximately three months after the end of radiotherapy. All patients were treated with adjuvant temozolomide [26]. More details about the patient population and treatment is provided by Seidlitz et al. [26]. Patients were excluded if no baseline MRI or no follow-up MRI was available, or if tumour or oedema extended across both hemispheres. Follow-up MRIs that showed motion artefacts or abnormal tissue in the normal appearing hemisphere were excluded, resulting in a total of 52 eligible patient datasets.
MRI data from six healthy controls (HCs) [age 39.4y ± 8.7 years, range (30.6–54.2 years), 4 male] were used to validate the data analysis strategy described in Sect. 2.3. This study was approved by the local ethics committee, and the participants gave written informed consent [DRKS-ID: DRKS00012600, EK267072017].
Data acquisition
All MRI data were acquired on a 3 T Philips Ingenuity PET/MR scanner (Philips, Eindhoven, The Netherlands) using an eight channel head coil. For this study, pre-contrast T1-weighted (T1w) MR images were used for further analysis. In patients, T1w images were acquired in sagittal orientation using a 3D turbo field echo (TFE) sequence at 1 mm isotropic resolution (TFE inversion prepulse, TFE factor = 224, TR/TE = shortest (typical 8.2/3.8 ms), α = 8°, FOV 192(FH) × 224(AP) mm2, matrix size 192(FH) × 224(AP), 1 mm slice thickness, 192 slices). In healthy controls, T1w images were acquired using a 3D gradient spoiled echo sequence in sagittal orientation (TR = 10 ms, TE = 3.7 ms, α = 20°, FOV 224 × 224 mm2, matrix size 224 × 224, 1 mm slice thickness, 160 slices) [27].
Data analysis
T1w images were segmented into GM, WM, CSF, bone, soft tissue and background using SPM12 [20] and GM, WM and CSF using FSL [18] with their respective default settings. SPM gave tissue probability maps as outputs, whereas FSL provided partial volume maps. For FSL, prior steps to segmentation included N4 bias field correction [28] and skull stripping [29]. The 3 T T1w images show relatively large bias field variations and without prior N4 bias field correction, FSL falsely segmented most of the subcortical GM as WM.
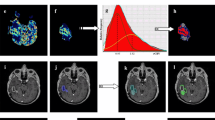
GM and WM volumes of the normal appearing hemisphere were compared between segmenting the whole brain (VGM,full, VWM,full, VGM + WM,full) versus segmenting the normal appearing hemisphere (VGM,cl, VWM,cl, VGM + WM,cl). The tumour-bearing and normal appearing hemispheres were separated by non-linearly coregistering the MNI152 atlas (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) to each T1w image using ANTs [29, 30]. The same deformation was applied to a label mask separating the two hemispheres. Subsequently, all GM and WM maps were restricted to the normal appearing hemisphere using these masks. GM and WM volumes were generated by summing all GM and WM values within these final tissue segmentation maps. Relative tissue volumes were calculated as ratios to their corresponding baseline values. The data analysis workflow is shown in Fig. 1. Mean WM and GM radiation doses in the contralateral hemisphere were calculated from the corresponding dose maps using binarized WM and GM maps, respectively, using a threshold of 0.5. The mean doses were used to compare the dose dependence of volume changes between the segmentation methods.
Image processing workflow: registration (Reg) of MNI152 atlas and corresponding left/right label to T1w-native space and subsequent tissue segmentation (Seg) of the full brain or contralateral (cl) hemisphere for volume assessment. In this example, abnormal tissue in the left hemisphere was mainly classified as GM (red arrow). Volume assessment was restricted to the contralateral hemisphere in all cases
The comparison between the two segmentation methods (full and cl) relies on the assumption that segmenting a single hemisphere will yield the same results in that hemisphere as segmenting the whole brain, thus serving as “ground truth” for patient data with one healthy and one abnormal hemisphere. To test this assumption, the T1w images of six healthy controls were segmented. GM volumes of the left and right hemispheres obtained from whole brain segmentation were compared with those obtained from segmenting the left and right hemispheres separately.
Statistical analysis
Relative GM, WM and GM + WM volumes of the contralateral hemispheres were compared between whole brain segmentation (full) and contralateral hemisphere segmentation (cl) using paired t tests. Normality of the distributions was tested using a one-sample Kolmogorov–Smirnov test. Cohen’s d = μx − μy/σx–y was used to calculate the effect size.
Relative GM and WM volumes obtained from segmenting the whole brain (full) and the contralateral hemisphere (cl) were correlated with the corresponding mean dose in GM and WM of the contralateral hemisphere, respectively, using Pearson regression analysis.
Results
Demographics of the 52 eligible patients are given in Table 1.
GM volumes from the healthy controls show that both FSL and SPM are suitable for segmentation of a single hemisphere with very little deviation between single-hemisphere and full brain segmentation (see supplementary material section I).
The relative GM and WM volume changes determined using full brain segmentation and contralateral hemisphere segmentation are compared in Fig. 2. With SPM, relative GM volume loss was significantly higher when using the full brain segmentation approach (mean ± std: ΔVGM,full = − 3.1% ± 3.7%, ΔVGM,cl = − 1.6% ± 2.7%; p < 0.001, d = 0.62). There was no significant difference in relative GM volume changes between full and cl segmentation when using FSL (mean ± std: ΔVGM,full = − 0.9% ± 2.7%, ΔVGM,cl = − 0.9% ± 2.5%; p = 0.91, d = 0.02). There was no significant difference in relative WM volume changes for SPM (mean ± std: ΔVWM,full = − 0.8% ± 2.4%, ΔVWM,cl = − 0.8% ± 2.1%; p = 0.95, d = 0.01) and FSL (mean ± std: ΔVWM,full = − 0.75% ± 2.4%, ΔVWM,cl = − -0.77% ± 2.2%; p = 0.89, d = 0.02).
Relative volume changes of grey matter (GM), white matter (WM) and the combination of the two volumes (GM + WM) between baseline and the 3 month follow-up for SPM and FSL. Results of full brain segmentation (full) and contralateral segmentation (cl) were compared using a paired t test (*p < 0.05). ΔV change in volume
When segmenting the full brain with SPM, relative GM volume changes (Fig. 3a) were not significantly correlated with the mean GM dose (r = − 0.23, p = 0.1). Additionally, a non-zero x = 0 intercept suggested GM atrophy despite a mean GM dose of 0 Gy. On the other hand, using the SPM cl, FSL full, and FSL cl segmentation, the relative GM volume changes (Fig. 3b, d, e) were significantly correlated with the mean GM dose (SPM cl: r = − 0.4, p = 0.004, FSL full: r = − 0.4, p = 0.004, FSL cl: r = − 0.35, p = 0.012) and the near zero intercepts suggest no GM changes with a mean GM dose of 0 Gy. The slopes of these three regressions were very similar, corresponding to a 1.1% (SPM cl), 1.1% (FSL full) and 0.9% (FSL cl) GM volume loss per 10 Gy mean GM dose.
Scatter plots of relative grey matter (GM) volume changes over the mean GM dose. GM volumes were determined from SPM and FSL segmentation of full brain (a, d) and contralateral hemisphere (b, e). Regression parameters were determined using Pearson’s correlation. The volume changes determined from full brain and cl segmentation are compared in (c, f). Red markers highlight four individual examples which are shown in Fig. 4. ΔVGM change in GM volume
The GM volume changes of the SPM cl (GMcl) and SPM full (GMfull) segmentations are compared in Fig. 3c. While the two methods produce very similar results for some cases (e.g. highlighted example A), there are large differences for other cases (highlighted examples B, C and D). GM difference maps between SPM cl (GMcl) and SPM full (GMfull) segmentation are shown for these four examples in Fig. 4. Example A shows similar relative GM volumes between full and cl segmentation in Fig. 3c and relatively consistent GMcl–GMfull difference maps (Fig. 4a). Conversely, examples B, C and D show the largest relative GM volume changes between full and cl segmentation in Fig. 3c. GM difference maps GMc–GMfull reveal these changes to be located at the GM/CSF interface around the gyri in example B (Fig. 4b), around the WM/GM interface in example C (Fig. 4c) and around both the GM/CSF and WM/GM interfaces in example D (Fig. 4d). All three examples B, C and D show pronounced MRI changes in abnormal tissue volume and contrasts between the baseline and follow-up T1w images in the ipsilateral hemispheres. Such outliers were not observed when using the FSL segmentation (Fig. 3f).
SPM segmentation: Differences of the grey matter (GM) probability maps of segmenting the contralateral hemisphere (GMcl, blue) minus segmenting the whole brain (GMfull, red). Examples a–d are highlighted in Fig. 3. FU follow-up
WM volume changes after 3 months were much smaller (supplementary Fig. 3), nevertheless, as for GM, WM atrophy was significantly correlated to the mean WM dose when using WM volumes obtained from SPM cl (r = − 0.28, p = 0.044), FSL full (r = − 0.29, p = 0.04) and FSL cl (r = − 0.41, p = 0.003) segmentation. The estimated degree of WM atrophy was 0.5% (SPM cl), 0.6% (FSL full) and 0.8% (FSL cl) per 10 Gy mean WM dose.
Discussion
We have shown that GM volume measurements in the contralateral hemisphere of patients with unilateral glioblastoma differ between segmentation of the whole brain and of the contralateral hemisphere alone when using SPM. This difference is likely caused by abnormal tissue changes in the ipsilateral hemisphere, typically seen during longitudinal observations of brain tumour patients, influencing normal tissue segmentation results with SPM in the contralateral hemisphere.
In the data presented in this study, higher GM atrophy was observed when not masking the ipsilateral hemisphere containing the abnormal tissue prior to SPM segmentation (Fig. 2). Since the WM volume shows no significant change, we conclude that the remaining volume is mainly assigned to CSF. Nevertheless, there were also examples in Fig. 4c, d where underestimated GM from the SPM full segmentation was also assigned to WM. Examples B, C and D have particularly large deviations in GM volume changes between full and cl SPM segmentation (Fig. 3c) and have also shown particularly pronounced image changes in abnormal tissue volume and contrast. It is therefore likely that such large regions of MRI changes will cause greater alterations to the normal tissue probability density distributions within the iterative segmentation process and thus shift tissue separation boundaries accordingly. An additional factor is that SPM also performs simultaneous bias field correction with the tissue segmentation by default [18, 20]. Depending on regularization strength of the bias field, this will also attempt to match abnormal tissue image intensities to normal tissue intensities [31]. Therefore, the overall impact of abnormal tissue on the segmentation depends on the interaction of a number of factors and is not easily appointed to a single cause. Nevertheless, we can conclude that abnormal tissue present in the ipsilateral hemisphere of brain tumour patients will influence SPM segmentation results of the contralateral hemisphere.
Conversely, FSL segmentation results are consistent between full brain and cl segmentation. Corresponding atrophy estimates are also very similar to those obtained from SPM cl segmentation for both GM (Fig. 3) and WM (suppl. Fig. 3). Unlike SPM, by default FSL uses grey value thresholds to coarsely segment the input image into the three expected tissue types GM, WM and CSF [18]. Although the abnormal tissue has to be assigned to one of those normal tissue types as with the SPM TPM method, this seems to have no apparent impact on the segmentation results. The weakness of the threshold-based initialisation of FSL are large bias field variations. These can cause the intensity values between GM, WM and CSF to overlap [18] and that led to misclassification of subcortical GM as WM in the data presented here. However, using the established N4 bias field correction algorithm [28] prior to FSL segmentation resulted in a much improved segmentation with visually very similar results to SPM.
The findings presented here are relevant for longitudinal studies requiring accurate brain volume measurement using iterative segmentation software with TPMs for initialization. While some previous studies have considered that abnormal tissue could bias volume measurements and restricted the segmentation to the contralateral hemisphere [4, 5], others have not [7, 32, 33]. Consequently, previous findings of volume changes determined with SPM could potentially be biased [32].
A limitation of this study is the restriction of the analysis to unilateral brain tumour cases, as done in some previous studies [4, 5]. However, by co-registering individual abnormality masks for all time points of a patient into a common space, arbitrary anatomical regions can be masked as demonstrated previously [27]. This allows accurate segmentation of normal appearing tissue in cases with bilateral lesions as well, albeit spatial normalization can be difficult in patients with large mass effect [34]. Minor differences in the FSL and SPM results could be caused by the fact that SPM gave tissue probability maps as outputs, whereas FSL provided partial volume maps. However, a preliminary analysis using tissue probability maps output for FSL (“-p” option) showed no change in the overall results. Lastly, the behaviour of other segmentation tools using a similar methodology, such as AFNI [35] or Atropos [36], was not tested. However, for accurate volumes measurements, we generally suggest to exclude abnormal tissue prior to segmentation in future studies.
In conclusion, abnormal tissue present in the ipsilateral hemisphere of brain tumour patients will influence SPM segmentation results of the contralateral hemisphere. In the dataset presented here, GM atrophy was overestimated in the contralateral hemisphere when the ipsilateral hemisphere containing abnormal tissue was not removed prior to segmentation. Consequently, for accurate volume measurement in brain tumour patients using SPM, the abnormal tissue needs to be masked prior to segmentation, however, this is not necessary when using FSL.
References
Ribi K, Relly C, Landolt MA, Alber FD, Boltshauser E, Grotzer MA (2005) Outcome of medulloblastoma in children: long-term complications and quality of life. Neuropediatrics 36:357–365
Greene-Schloesser D, Robbins ME (2012) Radiation-induced cognitive impairment–from bench to bedside. Neuro Oncol 14(Suppl 4):iv37–iv44
Makale MT, McDonald CR, Hattangadi-Gluth JA, Kesari S (2017) Mechanisms of radiotherapy-associated cognitive disability in patients with brain tumours. Nat Rev Neurol 13:52–64
Cayuela N, Jaramillo-Jiménez E, Càmara E, Majós C, Vidal N, Lucas A, Gil-Gil M, Graus F, Bruna J, Simó M (2019) Cognitive and brain structural changes in long-term oligodendroglial tumor survivors. Neuro Oncol 21:1470–1479
Petr J, Platzek I, Hofheinz F, Mutsaerts HJMM, Asllani I, van Osch MJP, Seidlitz A, Krukowski P, Gommlich A, Beuthien-Baumann B, Jentsch C, Maus J, Troost EGC, Baumann M, Krause M, van den Hoff J (2018) Photon vs. proton radiochemotherapy: effects on brain tissue volume and perfusion. Radiother Oncol 128:121–127
Karunamuni R, Bartsch H, White NS, Moiseenko V, Carmona R, Marshall DC, Seibert TM, McDonald CR, Farid N, Krishnan A, Kuperman J, Mell L, Brewer JB, Dale AM, Hattangadi-Gluth JA (2016) Dose-dependent cortical thinning after partial brain irradiation in high-grade glioma. Int J Radiat Oncol Biol Phys 94:297–304
Prust MJ, Jafari-Khouzani K, Kalpathy-Cramer J, Polaskova P, Batchelor TT, Gerstner ER, Dietrich J (2015) Standard chemoradiation for glioblastoma results in progressive brain volume loss. Neurology 85:683–691
Seibert TM, Karunamuni R, Bartsch H, Kaifi S, Krishnan AP, Dalia Y, Burkeen J, Murzin V, Moiseenko V, Kuperman J, White NS, Brewer JB, Farid N, McDonald CR, Hattangadi-Gluth JA (2017) Radiation dose-dependent hippocampal atrophy detected with longitudinal volumetric magnetic resonance imaging. Int J Radiat Oncol Biol Phys 97:263–269
Shi L, Feng-Lei Du, Sun Z-W, Zhang L, Chen Y-Y, Xie T-M, Li P-J, Huang S, Dong B-Q, Zhang M-M (2018) Radiation-induced gray matter atrophy in patients with nasopharyngeal carcinoma after intensity modulated radiotherapy: a mri magnetic resonance imaging voxel-based morphometry study. Quant Imaging Med Surg 8:902–909
Takeshita Y, Watanabe K, Kakeda S, Hamamura T, Sugimoto K, Masaki H, Ueda I, Igata N, Ohguri T, Korogi Y (2020) Early volume reduction of the hippocampus after whole-brain radiation therapy: an automated brain structure segmentation study. Jpn J Radiol 38:118–125
Lv X, He H, Yang Y, Han L, Guo Z, Chen H, Li J, Qiu Y, Xie C (2019) Radiation-induced hippocampal atrophy in patients with nasopharyngeal carcinoma early after radiotherapy: a longitudinal mr-based hippocampal subfield analysis. Brain Imaging Behav 13:1160–1171
Huynh-Le M-P, Karunamuni R, Moiseenko V, Farid N, McDonald CR, Hattangadi-Gluth JA, Seibert TM (2019) Dose-dependent atrophy of the amygdala after radiotherapy. Radiother Oncol 136:44–49
Raschke F, Seidlitz A, Wesemann T, Löck S, Jentsch C, Platzek I, Petr J, van den Hoff J, Kotzerke J, Beuthien-Baumann B, Baumann M, Linn J, Krause M, Troost EGC (2020) Dose dependent cerebellar atrophy in glioma patients after radio(chemo)therapy. Radiother Oncol 150:262–267
Nagtegaal SHJ, David S, van der Boog ATJ, Leemans A, Verhoeff JJC (2019) Changes in cortical thickness and volume after cranial radiation treatment: A systematic review. Radiother Oncol 135:33–42
Douw L, Klein M, Fagel SS, van den Heuvel J, Taphoorn MJ, Aaronson NK, Postma TJ, Peter Vandertop W, Mooij JJ, Boerman RH, Beute GN, Sluimer JD, Slotman BJ, Reijneveld JC, Heimans JJ (2009) Cognitive and radiological effects of radiotherapy in patients with low-grade glioma: long-term follow-up. Lancet Neurol 8:810–818
Fox NC, Scahill RI, Crum WR, Rossor MN (1999) Correlation between rates of brain atrophy and cognitive decline in ad. Neurology 52:1687–1689
Arnold DL, Li D, Hohol M, Chakraborty S, Chankowsky J, Alikhani K, Duquette P, Bhan V, Montanera W, Rabinovitch H, Morrish W, Vandorpe R, Guilbert F, Traboulsee A, Kremenchutzky M (2015) Evolving role of mri in optimizing the treatment of multiple sclerosis: Canadian consensus recommendations. Mult Scler J Exp Transl Clin 1:2055217315589775
Zhang Y, Brady M, Smith S (2001) Segmentation of brain mr images through a hidden markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20:45–57
Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, Nicola De Stefano J, Brady M, Matthews PM (2004) Advances in functional and structural mr image analysis and implementation as fsl. Neuroimage 23(Suppl 1):S208–S219
Ashburner J, Friston KJ (2005) Unified segmentation. Neuroimage 26:839–851
Ashburner J (2012) Spm: a history. Neuroimage 62:791–800
Guo C, Ferreira D, Fink K, Westman E, Granberg T (2019) Repeatability and reproducibility of freesurfer, fsl-sienax and spm brain volumetric measurements and the effect of lesion filling in multiple sclerosis. Eur Radiol 29:1355–1364
Vrenken H, Jenkinson M, Horsfield MA, Battaglini M, van Schijndel RA, Rostrup E, Geurts JJG, Fisher E, Zijdenbos A, Ashburner J, Miller DH, Filippi M, Fazekas F, Rovaris M, Rovira A, Barkhof F, de Stefano N, MAGNIMS Study Group (2013) Recommendations to improve imaging and analysis of brain lesion load and atrophy in longitudinal studies of multiple sclerosis. J Neurol 260:2458–2471
Selvaganesan K, Whitehead E, DeAlwis PM, Schindler MK, Inati S, Saad ZS, Ohayon JE, Cortese ICM, Smith B, Jacobson S, Nath A, Reich DS, Inati S, Nair G (2019) Robust, atlas-free, automatic segmentation of brain mri in health and disease. Heliyon 5:e01226
Valverde S, Oliver A, Roura E, Pareto D, Vilanova JC, Ramió-Torrentà L, Sastre-Garriga J, Montalban X, Rovira À, Lladó X (2015) Quantifying brain tissue volume in multiple sclerosis with automated lesion segmentation and filling. Neuroimage Clin 9:640–647
Seidlitz A, Beuthien-Baumann B, Löck S, Jentsch C, Platzek I, Zöphel K, Linge A, Kotzerke J, Petr J, van den Hoff J, Steinbach J, Krex D, Schmitz-Schackert G, Falk M, Baumann M, Krause M (2021) Final results of the prospective biomarker trial petra: 11c-met-accumulation in postoperative pet/mri predicts outcome after radiochemotherapy in glioblastoma. Clin Cancer Res 27:1351–136
Raschke F, Wesemann T, Wahl H, Appold S, Krause M, Linn J, Troost EGC (2019) Reduced diffusion in normal appearing white matter of glioma patients following radio(chemo)therapy. Radiother Oncol 140:110–115
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4itk: improved n3 bias correction. IEEE Trans Med Imaging 29:1310–1320
Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC (2011) A reproducible evaluation of ants similarity metric performance in brain image registration. Neuroimage 54:2033–2044
Tustison NJ, Avants BB (2013) Explicit b-spline regularization in diffeomorphic image registration. Front Neuroinform 7:39
Volz S, Nöth U, Jurcoane A, Ziemann U, Hattingen E, Deichmann R (2012) Quantitative proton density mapping: correcting the receiver sensitivity bias via pseudo proton densities. Neuroimage 63(1):540–552
Gommlich A, Raschke F, Wahl H, Troost EGC (2018) Retrospective assessment of mri-based volumetric changes of normal tissues in glioma patients following radio(chemo)therapy. Clin Transl Radiat Oncol 8:17–21
Riggs L, Bouffet E, Laughlin S, Laperriere N, Liu F, Skocic J, Scantlebury N, Wang F, Schoenhoff NJ, Strother D, Hukin J, Fryer C, McConnell D, Mabbott DJ (2014) Changes to memory structures in children treated for posterior fossa tumors. J Int Neuropsychol Soc 20:168–180
Visser M, Petr J, Müller DMJ, Eijgelaar RS, Hendriks EJ, Witte M, Barkhof F, van Herk M, Mutsaerts HJMM, Vrenken H, de Munck JC, De Witt Hamer PC (2020) Accurate mr image registration to anatomical reference space for diffuse glioma. Front Neurosci 14:585
Vovk A, Cox RW, Stare J, Suput D, Saad ZS (2011) Segmentation priors from local image properties: without using bias field correction, location-based templates, or registration. Neuroimage 55:142–152
Avants BB, Tustison NJ, Jue W, Cook PA, Gee JC (2011) An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics 9:381–400
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
In the past 5 years, Dr. Michael Baumann received funding for his research projects and for educational grants to the University of Dresden by Bayer AG (2016–2018), Merck KGaA (2014-open) and Medipan GmbH (2014–2018). He is on the supervisory board of HI-STEM gGmbH (Heidelberg) for the German Cancer Research Center (DKFZ, Heidelberg) and also member of the supervisory body of the Charité University Hospital, Berlin. As former chair of OncoRay (Dresden) and present CEO and Scientific Chair of the German Cancer Research Center (DKFZ, Heidelberg), he has been or is responsible for collaborations with a multitude of companies and institutions, worldwide. In this capacity, he has discussed potential projects and signed contracts for research funding and/or collaborations with industry and academia for his institute(s) and staff, including but not limited to pharmaceutical companies such as Bayer, Boehringer Ingelheim, Bosch, Roche and other companies such as Siemens, IBA, Varian, Elekta, Bruker, etc. In this role, he was/is also responsible for the commercial technology transfer activities of his institute(s), including the creation of start-ups and licensing. This includes the DKFZ-PSMA617 related patent portfolio [WO2015055318 (A1), ANTIGEN (PSMA)] and similar IP portfolios. Dr. Baumann confirms that, to the best of his knowledge, none of the above funding sources were involved in the preparation of this paper. Within the past years, Dr. Krause received funding for her research projects by IBA (2016), Merck KGaA (2014–2018 for preclinical study; 2018–2020 for clinical study), Medipan GmbH (2014–2018), Attomol GmbH (2019–2021), GA Generic Assays GmbH (2019–2021), BTU Cottbus-Senftenberg (2019–2021), Gesellschaft für medizinische und wissenschaftliche genetische Analysen (2019–2021), Lipotype GmbH (2019–2021), PolyAn GmbH (2019–2021). In the past 5 years, Dr. Troost has been involved in an ongoing publicly funded (German Federal Ministry of Education and Research) project with the companies Medipan, Attomol GmbH, GA Generic Assays GmbH, Gesellschaft für medizinische und wissenschaftliche genetische Analysen, Lipotype GmbH and PolyAn GmbH (2019–2021). Dr. Baumann, Dr. Krause and Dr. Troost confirm that to the best of their knowledge none of the above funding sources were involved in the preparation of this paper. The other authors have nothing to disclose.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gommlich, A., Raschke, F., Petr, J. et al. Overestimation of grey matter atrophy in glioblastoma patients following radio(chemo)therapy. Magn Reson Mater Phy 35, 145–152 (2022). https://doi.org/10.1007/s10334-021-00922-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-021-00922-3