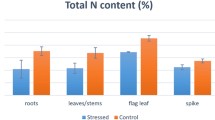
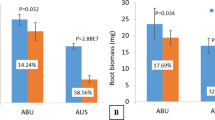
Abstract
Nitrogen is one of the essential elements for plant growth. Wild soybeans (Glycine soja) have strong abilities to survive in harsh and barren environments, and hence become ideal plant model for studying plant adaptability to low nitrogen (LN) conditions. In this study, we analyzed and compared the transcriptomes of wild soybean subjected to LN treatments. We totally identified 1095 (681 up and 414 down) and 5490 (2998 up and 2492 down) differentially expressed genes (DEGs) in the aerial parts (leaf and stem, LS) and roots, respectively. Gene ontology classification analysis revealed that the categories related to LN stress (including oxidation reduction, transcriptional regulation, membrane, and protein phosphorylation) were highly enriched among DEGs. In addition, a total of 784 transcription factor (TF) and 84 transporter protein (TP) genes were determined in LS DEGs, of which some TF genes (NAC1, NAC35, ZFP1, CIM1, and WRKY25) and TP genes like NRT2.5 (nitrate transporter) and ABCC12 (ABC transporter) were widely upregulated under LN stress. Nevertheless, a total of 3859 TF and 370 TP genes were identified in root DEGs, of which some TF genes (NAC6, NAC14, MYB29, MYB92, bZIP62, bZIP72, WRKY60, WRKY58) and TP genes like NRT2.4 and HAK5 (potassium transporter) were upregulated under LN stress. These findings suggest that the identified DEGs may play vital roles in plant responses to LN stress, providing important genetic resources for further functional dissection of plant molecular mechanisms to LN stress.












Similar content being viewed by others
Abbreviations
- CK:
-
control
- LS:
-
leaf and stem
- LN:
-
low nitrogen
- DEG:
-
differentially expressed gene
- TP:
-
transporter protein
- TF:
-
transcription factor
- NRT:
-
nitrate transporter
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Bräutigam A, Gowik U (2010) What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biology 12:831–841
Chen S, Jiang J, Li HY, Liu GF (2012) The salt-responsive transcriptome of Populus simonii × Populus nigra via DGE. Gene 504:203–212
Drew MC, Saker LR, Ashely TW (1973) Nutrient supply and the growth of the seminal root system inbarely. J Exp Bot 24(6):1189–1202
DuanMu HZ, Wang Y, Bai X, Cheng SF, Deyholos MK, Wong GK-S, Zhu D, Li R, Yu Y, Cao L, Chen C, Zhu YM (2015) Wild soybean roots depend on specific transcription factors and oxidation reduction related genesin response to alkaline stress. Funct Integr Genomic 15:651–660
Elbourne LD, Tetu SG, Hassan KA, Paulsen IT (2017) TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res 45:320–324
Gao QS, Zhang D, Xu L, Xu CW (2011) Systematic identification of rice ABC1 gene family and its response to abiotic stress. Rice Sci 18(3):167–177
Ge Y, Li Y, Zhu YM, Bai X, Lv DK, Guo DJ, Ji W, Cai H (2010) Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol 10(1):153
Gómez I, Pedreño JN, Moral R, Iborra MR, Palacios G, Mataix J (1996) Salinity and nitrogen fertilization affecting the macronutrient content and yield of sweet pepper plants. J Plant Nutr 19(2):353–359
Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36(10):3420–3435
Hikosaka K (2004) Interspecific difference in the photosynthesis–nitrogen relationship: patterns, physiological causes, and ecological importance. J Plant Res 117:481–494
Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, Zhang QF, Xiong LZ (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. PNAS 103:2987–2992
Jensen LJ, Bork P (2010) Ontologies in quantitative biology: a basis for comparison, integration, and discovery. PLoS Biol 8(5):e1000374
Jin JP, Tian F, Yang DC, Meng YQ, Kong L, Luo JC, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45:1040–1045
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:480–484
Kim DY, Bovet L, Kushnir S, Noh EW, Martinoia E, Lee Y (2006) AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol 140(3):922–932
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, Phillippy AM (2012) Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30(7):693–700
Larsen PB, Cancel J, Rounds M, Ochoa V (2007) Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225(6):1447–1458
Li Q, Ding GD, Yang NM, White PJ, Ye XS, Cai HM, Lu JW, Shi L, Xu F (2019) Comparative genome and transcriptome analysis unravels key factors of nitrogen use efficiency in Brassica napus L. Plant Cell Environ 43:712–731
Liu JT, Zhou YL, Luo CX, Xiang Y, An LZ (2016) De novo transcriptome sequencing of desert herbaceous Achnatherum splendens (Achnatherum) seedlings and identification of salt tolerance genes. Genes-Basel 7(4):1–13
Liu SY, Fukumoto T, Gena P, Feng P, Sun Q, Li Q, Matsumoto T, Kaneko T, Zhang H, Zhang Y, Zhong SH, Zeng WZ, Katsuhara M, Kitagawa Y, Wang AX, Calamita G, Ding XD (2020a) Ectopic expression of a rice plasma membrane intrinsic protein (OsPIP1;3) promotes plant growth and water uptake. Plant J 102:779–796
Liu DP, Li MX, Liu Y, Shi LX (2020b) Integration of the metabolome and transcriptome reveals the resistance mechanism to low nitrogen in wild soybean seedling roots. Environ Exp Bot 175
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆Ct method. Methods 25(4):402–408
Mao XZ, Cai T, Olyarchuk JG, Wei LP (2005) Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Nacry P, Bouguyon E, Gojon A (2013) Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 370:1–29
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozali K (2007) Function analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295
Rahman H, Jagadeeshselvam N, Valarmathi R, Sasikala R, Senthil N, Sudhakar D, Robin S, Muthurajan R (2014) Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Plant Mol Biol 85(4-5):485–503
Schuster SC (2008) Next-generation sequencing transforms today's biology. Nat Methods 5(1):16
Shortemeyer M, Feil B, Stamp P (1993) Root morphology and nitrogen uptake of maize simultaneously supplied with ammonium and nitrate in a split-root system. Ann Bot-London 72(2):107–115
Sinha SK, Sevanthi VA, Chaudhary S, Tyagi P, Venkadesan S, Rani M, Mandal P (2018) Transcriptome Analysis of Two Rice Varieties Contrasting for Nitrogen Use Efficiency under Chronic N Starvation Reveals Differences in Chloroplast and Starch Metabolism-Related Genes. Genes 9(4):206
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–U174
Wu GS, Carville JS, Spalding EP (2016) ABCB19-mediated polar auxin transport modulates Arabidopsis hypocotyl elongation and the endoreplication variant of the cell cycle. Plant J 85(2):209–218
Wu B, Hu YN, Huo PJ, Zhang Q, Chen X, Zhang ZW (2017) Transcriptome analysis of hexaploid hulless Oat in response to salinity stress. PLoS One 12(2):e0171451
Xu ZC, Peters RJ, Weirather J, Luo HM, Liao BS, Zhang X, Zhu YJ, Ji AJ, Zhang B, Hu SN, An KF, Song JY, Chen SL (2015) Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J 82(6):951–961
Yadav SK (2010) Heavy metals toxicity in plants: an overview on the role of glutathione and phytochelatins in heavy metal stress tolerance of plants. S AFR J BOT 76:167–179
Yamaguchi T, Hamamoto S, Uozumi N (2013) Sodium transport system in plant cells. Front Plant Sci 4:410
Yang LF, Jin YH, Huang W, Sun Q, Liu F, Huang XZ (2018) Full-length transcriptome sequences of ephemeral plant Arabidopsis pumila provides insight into gene expression dynamics during continuous salt stress. BMC Genomics 19:717
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2):R14
Zhang LC, Zhao GY, Jia JZ, Liu X, Kong XY (2012) Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J Exp Bot 63:203–214
Zhang JY, Wang Y, Zhao YF, Zhang Y, Zhang JY, Ma HR, Han YZ (2020) Transcriptome analysis reveals Nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (Solanum tuberosum L.). PLoS ONE 15(10):e0240662
Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6:486–503
Zhou AM, Ma HP, Liu EH, Jiang TT, Feng S, Gong SF, Wang JG (2017) Transcriptome sequencing of Dianthus spiculifolius and analysis of the genes involved in responses to combined cold and drought stress. Int J Mol Sci 18:849
Zhou AM, Sun HW, Dai SY, Feng S, Zhang JZ, Gong SF, Wang JG (2019) Identification of transcription factors involved in the regulation of flowering in Adonis Amurensis through combined RNA-seq transcriptomics and iTRAQ proteomics. Genes 10(4):305
Acknowledgements
We thank the Lianchuan Biological Company (Hangzhou, China) for Illumina sequencing and raw data analysis.
Funding
This work was financially supported by National Natural Science Foundation of China (31670272 to X.D.), Natural Science Foundation of Heilongjiang Province (C2017014 to X.D.), and the Starting Fund of Northeast Agricultural University (to X.D.).
Author information
Authors and Affiliations
Contributions
QS drafted the manuscript. QS and HRL analyzed the data. HRL and QZ performed the phenotypic evaluation. DZ, JC, and JLX helped to coordinate the study. XDD and QL participated in the design of the study and revised the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable
Consent to participate
Not applicable
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by: Izabela Pawłowicz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(RAR 4759 kb)
Rights and permissions
About this article
Cite this article
Sun, Q., Lu, H., Zhang, Q. et al. Transcriptome sequencing of wild soybean revealed gene expression dynamics under low nitrogen stress. J Appl Genetics 62, 389–404 (2021). https://doi.org/10.1007/s13353-021-00628-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-021-00628-1