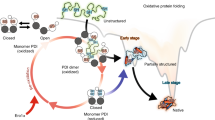
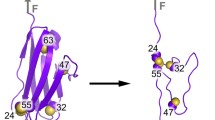
Abstract
The folding of proteins that contain disulfide bonds is termed oxidative protein folding. It involves a chemical reaction resulting in the formation of disulfide bonds and a physical conformational folding reaction that promotes the formation of the native structure. While the presence of disulfide bonds significantly increases the complexity of the folding landscape, it is generally recognized that native disulfide bonds help funnel the trajectory towards the final folded form. Here, we review the role of disulfide bonds in oxidative protein folding and argue that even structure-inducing native disulfide bond formation treads a fine line in the regeneration of disulfide-bond-containing proteins. The translation of this observation to protein misfolding related disorders is discussed.




Similar content being viewed by others
References
Gilbert HF (1990) Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol 63:69–172. https://doi.org/10.1002/9780470123096.ch2
Narayan M, Welker E, Wedemeyer WJ, Scheraga HA (2000) Oxidative folding of proteins. Acc Chem Res 33:805–812
Creighton TE (1997) Protein folding coupled to disulphide bond formation. Biol Chem 378:731–744
Wedemeyer WJ, Scheraga HA (2001) Protein folding: overview of pathways. Encyclopedia of life sciences. Wiley, Chichester
Welker E, Narayan M, Wedemeyer WJ, Scheraga HA (2001) Structural determinants of oxidative folding in proteins. Proc Natl Acad Sci USA 98:2312–2316
Wedemeyer WJ, Welker E, Narayan M, Scheraga HA (2000) Disulfide bonds and protein folding. Biochemistry 39:4207–4216
Welker E, Wedemeyer WJ, Narayan M, Scheraga HA (2001) Coupling of conformational folding and disulfide-bond reactions in oxidative folding of proteins. Biochemistry 40:9059–9064
Tu BP, Weissman JS (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences: mechanisms and consequences. J Cell Biol 164:341–346
Hudson DA, Gannon SA, Thorpe C (2015) Oxidative protein folding: from thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum. Free Radic Biol Med 80:171–182
Woycechowsky KJ, Raines RT (2000) Native disulfide bond formation in proteins. Curr Opin Chem Biol 4:533–539
Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J (1993) A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA 90:1038–1042
Mamathambika BS, Bardwell JC (2008) Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol 24:211–235
Rothwarf DM, Scheraga HA (1993) Regeneration of bovine pancreatic ribonuclease A. 1. Steady-state distribution. Biochemistry 32:2671–2679
Rothwarf DM, Scheraga HA (1993) Regeneration of bovine pancreatic ribonuclease A. 2. Kinetics of regeneration. Biochemistry 32:2680–2689
Rothwarf DM, Scheraga HA (1993) Regeneration of bovine pancreatic ribonuclease A. 3. Dependence on the nature of the redox reagent. Biochemistry 32:2690–2697
Rothwarf DM, Scheraga HA (1993) Regeneration of bovine pancreatic ribonuclease A. 4. Temperature dependence of the regeneration rate. Biochemistry 32:2698–2703
Rothwarf DM, Li YJ, Scheraga HA (1998) Regeneration of bovine pancreatic ribonuclease A: identification of two nativelike three-disulfide intermediates involved in separate pathways. Biochemistry 37:3760–3766
Rothwarf DM, Li YJ, Scheraga HA (1998) Regeneration of bovine pancreatic ribonuclease A: detailed kinetic analysis of two independent folding pathways. Biochemistry 37:3767–3776
Xu X, Rothwarf DM, Scheraga HA (1996) Nonrandom distribution of the one-disulfide intermediates in the regeneration of ribonuclease A. Biochemistry 35:6406–6417
Volles MJ, Xu X, Scheraga HA (1999) Distribution of disulfide bonds in the two-disulfide intermediates in the regeneration of bovine pancreatic ribonuclease A: further insights into the folding process. Biochemistry 38:7284–7293
Welker E, Narayan M, Volles MJ, Scheraga HA (1999) Two new structured intermediates in the oxidative folding of RNase A. FEBS Lett 460:477–479
Narayan M, Welker E, Scheraga HA (2001) Development of a novel method to study the rate-determining step during protein regeneration: application to the oxidative folding of RNase A at low temperature reveals BPTI-like kinetic traps. J Am Chem Soc 123:2909–2910
Weissman JS, Kim PS (1991) Reexamination of the folding of BPTI: predominance of native intermediates. Science 253:1386–1393
Creighton TE (1992) The disulfide folding pathway of BPTI. Science 256:111–114
Roux P, Ruoppolo M, Chaffotte AF, Goldberg ME (1999) Comparison of the kinetics of S-S bond, secondary structure, and active site formation during refolding of reduced denatured hen egg white lysozyme. Protein Sci 8:2751–2760
Patel AS, Lees WJ (2012) Oxidative folding of lysozyme with aromatic dithiols, and aliphatic and aromatic monothiols. Bioorg Med Chem 20:1020–1028
Arai K, Shibagaki W, Shinozaki R, Iwaoka M (2013) Reinvestigation of the oxidative folding pathways of hen egg white lysozyme: Switching of the major pathways by temperature control. Int J Mol Sci 14:13194–13212
Narayan M (2020) Revisiting the formation of a native disulfide bond: consequences for protein regeneration and beyond. Molecules 25:5337. https://doi.org/10.3390/molecules25225337
Chang JY, Ventura S (2011) Folding of disulfide proteins. Springer, Berlin/Heidelberg
Chakravarty S, Varadarajan R (2000) Elucidation of determinants of protein stability through genome sequence analysis. FEBS Lett 470:65–69
Patel S, Indu S, Ramakrishnan C, Varadarajan R (2013) Protein disulfide analysis and design. Biomolecular forms and functions. World Scientific/Indian Inst of Science, Bangalore, pp 296–311
Schulte L, Mao J, Reitz J, Sreeramulu S, Kudlinzki D, Hodirnau VV, Meier-Credo J, Saxena K, Buhr F, Langer JD et al (2020) Cysteine oxidation and disulfide formation in the ribosomal exit tunnel. Nat Commun 2020:11. https://doi.org/10.1038/s41467-020-19372-x
Mossuto MF (2013) Disulfide bonding in neurodegenerative misfolding diseases. Int J Cell Biol 2013:318319
Narayan M (2012) Disulfide bonds: protein folding and subcellular protein trafficking: disulfide bonds. FEBS J 279:2272–2282
Narayan M (2011) The case of oxidative folding of ribonuclease A: factors impacting fold maturation of ER-processed proteins. Folding of disulfide proteins. Springer, New York, pp 23–42
Cao P, Abedini A, Raleigh DP (2013) Aggregation of islet amyloid polypeptide: from physical chemistry to cell biology. Curr Opin Struct Biol 23:82–89
Acknowledgements
The author would like to thank Mr. Gyan M. Narayan for assistance with the References.
Funding
MN acknowledges support from NIH 1SC3 GM111200 01A1 for this work. The authors declare no competing financial interests.
Author information
Authors and Affiliations
Contributions
MN conceived the topic and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflict to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Narayan, M. The Formation of Native Disulfide Bonds: Treading a Fine Line in Protein Folding. Protein J 40, 134–139 (2021). https://doi.org/10.1007/s10930-021-09976-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-021-09976-7