Abstract
Background
Carotenoid biosynthesis is essential for the generation of photosynthetic pigments, phytohormone production, and flower color development. The light harvesting like 3 (LIL3) protein, which belongs to the light-harvesting complex protein family in photosystems, interacts with geranylgeranyl reductase (GGR) and protochlorophyllide oxidoreductase (POR) both of which are known to regulate terpenoid and chlorophyll biosynthesis, respectively, in both rice and Arabidopsis.
Results
In our study, a CRISPR-Cas9 generated 4-bp deletion mutant oslil3 showed aberrant chloroplast development, growth defects, low fertility rates and reduced pigment contents. A comparative transcriptomic analysis of oslil3 suggested that differentially expressed genes (DEGs) involved in photosynthesis, cell wall modification, primary and secondary metabolism are differentially regulated in the mutant. Protein-protein interaction assays indicated that LIL3 interacts with phytoene synthase (PSY) and in addition the gene expression of PSY genes are regulated by LIL3. Subcellular localization of LIL3 and PSY suggested that both are thylakoid membrane anchored proteins in the chloroplast. We suggest that LIL3 directly interacts with PSY to regulate carotenoid biosynthesis.
Conclusion
This study reveals a new role of LIL3 in regulating pigment biosynthesis through interaction with the rate limiting enzyme PSY in carotenoid biosynthesis in rice presenting it as a putative target for genetic manipulation of pigment biosynthesis pathways in crop plants.
Similar content being viewed by others
Introduction
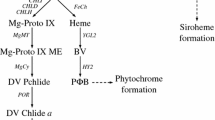
Light harvesting complex (LHC) proteins constitute major light harvesting antenna of photosynthetic eukaryotes. LHC proteins contain a representative LHC motif consisting of 25–30 amino acids (mostly hydrophobic). The LHC motif is also present in a number of transmembrane proteins such as light harvesting like (LIL) proteins (Takahashi et al. 2014). There are multiple LIL proteins in both photosynthetic eukaryotes and cyanobacteria. For example, there are five different types of LIL proteins in the model cyanobacterium Synechocystis sp. PCC6803 while there are at least eight different types of LIL proteins in Arabidopsis (Jansson 1999; Funk and Vermaas 1999; Liu et al. 2004). Among these LIL proteins, ferrochelatase (FC) and LIL3 have been functionally characterized. FC is responsible for the final step of Fe2+ insertion into protoporphyrin during heme biosynthesis (Funk and Vermaas 1999). LIL3 is involved in the stabilization of geranylgeranyl reductase (GGR), that is a key enzyme for phytyl pyrophosphate (Phytyl PP) formation (Tanaka et al. 2010).
In Arabidopsis, there are two LIL3 isoforms, namely AtLIL3.1 and AtLIL3.2, both of which share high sequence similarity (73% identity) with each other at the amino acid level. Both AtLIL3.1 and AtLIL3.2 not only regulate the stability of protochlorophyllide oxidoreductase (POR) enzymes through a direct physical interaction but also bind to protochlorophyllides, that are substrates of POR enzymes (Hey et al. 2017; Mork-Jansson and Eichacker 2018). Arabidopsis LIL3 also interacts with POR and GGR, thus modulating the levels of two metabolites, chlorophyllide and phytyl pyrophosphate, that are biosynthesized by POR and GGR1, respectively. These two metabolites are essential for chlorophyll and tocopherol synthesis in Arabidopsis (Hey et al. 2017). In rice, there is only one LIL3 ortholog and bi-molecular fluorescent complementation (BiFC) assay confirmed the interaction between LIL3 and GGR, similar to the observation in Arabidopsis (Zhou et al. 2017). OsLIL3 was also reported to interact with geranylgeranyl diphosphate synthase (OsGGPPS1) but not with OsGRP (OsGGPPS1-recruiting protein). OsGRP directs OsGGPPS1 to form a large multiprotein complex in thylakoid membrane consisting of OsGGR (geranylgeranyl reductase), OsGRP, OsCHLG (chlorophyll synthase) and OsPORB (protochlorophyllide oxidoreductase) (Zhou et al. 2017).
In rice, many leaf color mutants have been extensively studied providing insights into chlorophyll biosynthesis, chloroplast development, tetrapyrrole biosynthesis and photosynthesis (Nagata et al. 2005; Adhikari et al. 2011; Deng et al. 2014). One prominent phenotype of leaf color mutants is chlorotic lesion.
Apart from being deficient in photosynthetic pigments due to the disruption of pigment biosynthesis enzyme coding gene, the mutant phenotype can be a result of various stress conditions such nitrate and iron deficiency that affects pigment production (Shao et al. 2020; Shim et al. 2020). Leaf chlorosis is commonly related to chlorophyll biosynthesis and degradation pathways. Thus, deficiency in any step leading to chlorophyll synthesis can possibly result in a reduced chlorophyll content and thereby changes in leaf color. The LIL3 protein associates with the GGR and POR enzymes in both rice and Arabidopsis. These enzymes are involved in the generation of phytylated GGPP, a metabolite essential for both terpenoid and chlorophyll biosynthesis (Zhou et al. 2017; Hey et al. 2017). Thus, LIL3 acts as a common player in both chlorophyll and carotenoid biosynthesis.
In this study, we found that oslil3 mutant has a chlorotic phenotype and aberrant chloroplast development at seedling stages. RNA-seq analysis of oslil3 mutant found that DEGs involved in photosynthesis, cell wall and metabolism are differentially regulated in this mutant and protein-protein interaction assays found that LIL3 interacts with PSY enzymes and this interaction possibly effects PSY gene expression, revealing a novel regulatory mechanism of LIL3 in the carotenoid biosynthesis. Interestingly, LIL3 co-localized with PSY in the thylakoid membrane complex. LIL3 was also found to regulate the transcript levels of other LIL3-interacting protein genes including OR, POR, and GGR as well as the protein abundance of POR and GGR enzymes that were repressed in the oslil3 mutant. This shows the dramatic effect of LIL3 mutation on plant pigment pathways and its possible use in molecular biotechnology for crop pigment level manipulation.
Results
OsLIL3 CRISPR-Cas9 Mutant Displays Leaf Chlorosis and Developmental Defects
CRISPR-Cas9 was used for generation of a rice oslil3 mutant, taking advantage of gene editing protocol (Shan et al. 2014). The single guide RNA, sgRNA 5′-GGCGCCTCGACAGTCTCCACGGG − 3′ was designed for positional targeting of OsLIL3. A total of eight independent transgenic lines were obtained and out of these, one CRISPR-Cas9 generated mutant line showed stable homozygous oslil3 mutation and would be hereby referred to as oslil3 (16-#1) or oslil3 (Supplementary data S4). The T2 generation oslil3 (16-#1) homozygous plants displayed chlorotic phenotype at normal growth temperatures (28 °C), more prominently at the early seedling stages (Fig. 1a) but this phenotype was less visible at later (adult) stages (Fig. 1b). Therefore, this line was used for all the downstream experiments.
Phenotypic characterization of oslil3 mutant
(a), Phenotype of 3 weeks old WT and oslil3 mutant at 28 °C. (b), Morphological phenotype of adult plants of WT and oslil3. (c), Determination of exact mutation site by DNA sequencing. The deleted bases were indicated with a red line. (d), Immunoblot analysis of previously identified OsLIL3-interacting proteins in oslil3 mutant. OsAtpB was used as an internal reference protein. (e), Pigment characterization of L4 stage WT and oslil3 16-#1 mutant. Triple-asterisks indicates P < 0.001 (Student’s t-test, n = 6), and values are mean ± SD). FW, fresh weight. (f), TEM-based structural analysis of chloroplasts of L4 leaf in 4-leaf seedlings of WT and oslil3 mutant grown at 28 °C.
Sequencing revealed that oslil3 mutation in this line was caused by the deletion of four base pairs from the first exon of OsLIL3 (Fig. 1c), leading to a frameshift in translation. Sequence alignment with WT sequence showed that a deletion of four bases (positions 158–161) in the OsLIL3 coding sequence (Figure S1-a), resulted in premature translational termination (Figure S1-b). We also examined the top five ranked off-target sites predicted by CRISPR-P (Liu et al. 2017), and off-target mutations were located only to introns or CDS of transposon genes (Supplementary data S1).
Interestingly, the pigment content of oslil3 was greatly reduced. The mutant showed reduction of 80% of total chlorophylls and 60% of carotenoids, in comparison to the WT plants (Fig. 1e). TEM examination of ultrastructure of third leaf (L3) chloroplasts in oslil3 and wild-type 3-leaf seedlings grown at 28 °C showed highly disorganized thylakoid lamellar structures in oslil3 chloroplasts and without grana formation (Fig. 1f).
To account for the pigment reduction in oslil3, the effect of OsLIL3 on the regulation of proteins involved in carotenoid biosynthesis (OsGGR and OsPORB, previously reported interactors of OsLIL3) was investigated. Firstly, the peptide “AMIGFFMAYFVDSL” (172–185 amino acids of OsLIL3 protein sequence, used for generating LIL3 antibody) was used for analyzing protein expression in oslil3 vs WT. The results indicated absence of OsLIL3 in the mutant (Fig. 1d). The immunoblot analysis of OsLIL3-interacting proteins suggested that OsPORB and OsGGR protein levels were also drastically reduced in oslil3 vs WT at the 3-leaf stage seedlings (Fig. 1d), consistent with the previous findings (Tanaka et al. 2010; Hey et al. 2017).
To analyze the growth and yield performance of oslil3 (16-#1), plants grown for 90 days were harvested and different growth parameters were measured. Upon maturity, the oslil3 plants displayed significant differences in various quantified morphological changes in comparison to WT (Table 1), including reductions in plant height, culm length, ear number, tiller number and fertility rate in the oslil3 mutant. Overall, oslil3 was dwarf and yellowish with low fertility, with the chlorotic phenotype being more prominent in the early seedling stages.
Metabolism, Cell Wall Modification and Plant-Pathogen Interaction Genes Are Largely Affected in oslil3
To understand the molecular changes that could lead to the chlorotic phenotype of oslil3 mutant at seedling stages, we utilised genome wide transcriptome quantification. Mutation of OsLIL3 led to drastic gene expression changes in light reaction process, phenylpropanoid biosynthesis, carbon metabolism, terpenoid and amino acid biosynthesis.
Principal component analysis (PCA) suggested close clustering of sample replicates suggesting the desired sample reproducibility in RNA-Seq (Figure S2-a). Furthermore, gene expression correlation analysis mirrored the interpretation obtained from PCA (Figure S2-b). Differential gene expression analysis suggested substantial transcriptome modulation in oslil3 (vs WT). In total, a number of 4288 differentially expressed genes (DEGs) were identified, with 2188 up-regulated and 2100 down-regulated (Fig. 2a).
Overall description of RNA-sequencing data. (a), Number of up-regulated and down-regulated DEGs identified in mutant vs WT. (b), Overview of transcript abundance changes in oslil3 and WT in MapMan assigned pathway genes. Each gene is represented as one colored square in the metabolic map, with red representing downregulation and green representing upregulation. The color scale is based on log2(fold change) values of genes
Gene Ontology (GO) and KEGG pathway enrichment analyses were carried out for 4288 DEGs to interpret the functional implications of OsLIL3 mutation (Figure S3 and S4). In the biological process category, enriched GO terms consisted of aldonate transmembrane transport, chitin metabolic and catabolic process and catabolic processes of glucosamine-containing compound, amino sugar, aminoglycan and cell wall macromolecule. In the molecular function category, L-proline transmembrane transporter, endochitinase, glycogen debranching, (−)-secoisolariciresinol dehydrogenase enzyme, anthocyanidin 3-O-glucosyltransferase, and chitin activities were the top enriched terms. In the cellular component category, chloroplast thylakoid lumen, plastid thylakoid lumen, photosynthesis II oxygen evolving complex, thylakoid lumen, and anchored component of plasma membrane were the topmost enriched terms.
For specific metabolic pathways, top 20 KEGG pathways involving most number of DEGs are listed (Figure S4). DEGs were mostly enriched in terms such as “plant-pathogen interaction”, “phenylpropanoid biosynthesis”, “plant hormone signal transduction”, “amino sugar and nucleotide sugar metabolism”, “biosynthesis of amino acids” among others. Interestingly, the KEGG pathway term “plant-pathogen interaction” contained the greatest number of DEGs (listed in Supplementary data S5). In total, there are 19 genes in this category, including those encoding calcium-dependent protein kinase (CPK), respiratory burst oxidase (RBOH), calcium-binding protein (CML), nitrite oxide synthase (NAO1), WRKY transcriptional factors (WRKY22 & WRKY33), pathogen related protein 1 (PR1), LRR receptor like kinase FLS2 (FLS2), LRR receptor like kinase EFR (EFR), pto-interacting protein 1 (PIT1), disease resistance protein 3 (RPS3), RPS2, RPS5, chitin elicitor-binding protein (CEBiP) and receptor kinase-like protein (XA21).
We mapped the DEGs in our transcriptome analysis to different functional categories in the MapMan tool to visualize the pathways mostly affected by OsLIL3 deficiency. Metabolic pathways in cell wall biosynthesis, light reactions, lipid metabolism, secondary metabolism, amino acid metabolism, tetrapyrroles biosynthesis, and nucleotide metabolism were mostly changed (Fig. 2b). Specifically, most of the light reaction genes are up-regulated and majority of cell wall related genes are down-regulated.
OsLIL3 Mutation Largely Perturbs Expression of LHC Protein-Encoding, Cell Wall Metabolism and Pigment Biosynthesis Genes
MapMan analysis above revealed that almost 30 genes involved in light reactions are up-regulated and four genes are down-regulated in oslil3 (Fig. 3a, Supplementary data S6). Among those up-regulated genes, there is one PsbP, two PsbP-like genes (PPL1 and PPL2), Psb27, Psb related gene (OEC and OEC related), two Fru-1,6-bisphosphatase (FBPase) genes, one phosphoglycerate kinase (PGK), 15 in photosystem II polypeptide subunit genes, one cytochrome b6/f (CCB3), four NADH-plastoquinone oxidoreductase subunit genes (Ndh complex), two genes of ferredoxin 3 (FD3), and one plastocyanin (DRT112). The four down-regulated genes are psbS gene, glycocate oxidase (GOX), glycine cleavage system H gene (GCSH) and rubisco small subunit (RBCS1). In addition, genes involved in the cell wall metabolism were significantly down-regulated (about 77 genes in total) (Fig. 3b, Supplementary data S7). For example, six fasciclin-like AGP (FLA) genes, three cellulose synthase-like D family genes (CLSD) and two cellulose synthase-like A (CLSA) genes, four cellulose synthase genes (CESA), six xyloglucan endotrans-glycosylase (known as XTR, XTH, XET), nine expansin (EP) genes and the rest of them are all cell wall synthesis related genes.
Illustration of gene expression in metabolic pathways transcriptionally affected in oslil3 vs WT. (a), Up regulation of genes involved in light reactions by MapMan bin system. (b), Downregulation of genes involved in cell wall metabolism (c), Gene expression in chlorophyll biosynthesis, carotenoid biosynthesis and gibberellin biosynthesis genes: CPOX, coproporphyrinogen III oxidase; CHLI, magnesium chelatase subunit I; POR, protochlorophyllide reductase; KSL5, ent-pimara-8(14),15-diene synthase; CP71Z6, ent-isokaurene C2-hydroxylase; KSL6, ent-isokaurene synthase; MAS, momilactone-A synthase; KAO, ent-kaurenoic acid hydroxylase; GGR, geranylgeranyl reductase; NECD, 9-cis-epoxycarotenoid dioxygenase; CPS4, syn-copalyl-diphosphate synthase; LUT1, carotene epsilon-monooxygenase; PSY, phytoene synthase. Enzyme involved in each step was represented in red color show down-regulated genes while green color depicts up-regulated genes
Previously, it was reported that LIL3 interacts with GGR and POR enzymes with following experimental evidences in the oslil3osggr double mutant: a) higher accumulation of chlorophylls with unsaturated geranylgeraniol side chains in isoprenoid and chlorophyll biosynthesis, b) suppressed chloroplast development and c) plant lethality in the double mutant at L3 stages (Li et al. 2019). In our dataset, total 13 genes involved in isoprenoid biosynthesis were affected with 8 up-regulated genes (OsCPOX, OsCHLI, OsLUT1, OsMAS, OsCSP4, OsKSL5, OsKAO, and OsGA2OX) and 5 down-regulated genes (OsPOR, OsNCED, OsGGR, OsKSL6, and OsCY71Z6) (Fig. 3b, Supplementary data S8). These genes are involved in the branching pathways of geranylgeranyl diphosphate (GGPP), as it is an intermediate common metabolite in the biosynthesis of chlorophyll, lutein, GA, tocopherol, and momilactone A. Consistently, the POR and GGR enzymes were significantly down-regulated in our dataset as was also reported in the previous studies (Zhou et al. 2017; Hey et al. 2017; Li et al. 2019).
OsLIL3 Interacts with OsPSY Enzymes Involved in Carotenoid Biosynthesis
In rice, there is a single copy gene of LIL3 protein, referred to as OsLIL3, that shares high amino acid similarities and conserved functional domains with LIL3 in Arabidopsis (Figure S6). In addition to GGR and POR, it would be intriguing to find if OsLIL3 may possibly have other novel interactors among the enymes of chlorophyll and carotenoid pathways that are also perturbed in the oslil3 transcriptome. Therefore, Y2H assays were carried out to predict interactions between OsLIL3 and proteins in carotenoid biosynthesis pathways whose genes were identified as being differentially expressed in oslil3. Results suggested that the enzyme phytoene synthase 2 (PSY2, Os12g43130) interacts with OsLIL3 in yeast (Fig. 4a). A co-immunoprecipitation assay in tobacco leaves confirmed an interaction between OsLIL3 and OsPSY2 in vivo (Fig. 4b). An in planta BiFC assay revealed strong yellow fluorescence signals with co-expression of OsLIL3-YFPN and OsPSY2-YFPC constructs (Fig. 4c, top panel). No yellow fluorescent signals were detected in empty YFPN vector + OsPSY-YFPC or in OsLIL3-YFPN + empty YFPC vector controls. There are two other ortholog genes in addition to OsPSY2 (OsPSY1 and OsPSY3), and both of them share 74% identity to OsPSY2 at amino acid levels. To investigate whether OsPSY1 and OsPSY3 can interact with OsLIL3, above methods (Y2H, Co-IP and BiFC) were employed. The data suggests that OsLIL3 can also interact with OsPSY1 and OsPSY3 (Figure S7). In addition, interaction of OsLIL3 with other pigment metabolism proteins (OsLUT1, OsNCED2, OsCHLI and OsCPOX) was tested but no interactions were detected for these protein pairs in the Y2H assay (Figure S8). Taken together, these results indicated that OsLIL3 interacts physically with all three OsPSYs (inside chloroplasts in planta and also in yeast), suggesting that OsLIL3 may regulate carotenoid biosynthesis via its interaction with OsPSY enzymes.
Protein-protein interaction between OsLIL3 and OsPSY2. (a), Yeast two hybrid assays. OsLIL3 and LHC domains were fused with Gal DNA-binding domain (BD). OsPSY2 was fused with Gal DNA-activation domain (AD). The two other combinations OsLIL3/empty (pGADT7 vector) and empty (pGBKT7 vector)/OsPSY2 were included as negative controls. Yeast cells containing various vector combinations were grown on DDO plates (SD –Leu –Trp) for co-transformation selection. Yeast cells showing growth and blue color on high selection TDO/X/A plates (SD-His-Leu-Trp + X-alfa-Gal + Aureobasidin A) confirmed positive interactions between the two indicated proteins. (b), OsLIL3-FLAG coimmunoprecipitated with OsPSY2-HA. Total protein was purified from tobacco leaves expressing OsLIL3-FLAG with or without OsPSY2-HA and immunoprecipitated with an anti-Flag or anti-HA antibody. The presence of OsLIL3-FLAG, and OsPSY2-HA in the complex was determined by immunoblot analysis with Flag anti-bodies and anti-HA anti-bodies respectively. (c), BiFC-visualized interactions between OsLIL3 and OsPSY2 in N. benthamiana leaves. Abbreviations: YN, YFPN. YC, YFPC. Chl, chlorophyll autofluorescence. BF, bright field. All bars = 5 μm
OsLIL3 and OsPSY2 Co-Localize to Thylakoid Membranes of Chloroplasts
Confocal microscope-based visualization of N. benthamiana leaves infiltrated with p35S-OsLIL3-EYFP and p35S-OsPSY2-EYFP revealed the localization of yellow fluorescence signals for both constructs to the chloroplasts of mesophyll cells (Fig. 5a).
Subcellular localization of OsLIL3 and OsPSY2 in N. benthamiana leaves
Fluorescent signals of OsLIL3-EYFP and OsPSY2-EYFP indicated by confocal microscopy. In both cases, chlorophyll autofluorescence (Chl) merged with yellow fluorescence signals, suggesting chloroplast localization. All bars = 10 μm. The OsLIL3-EYFP and OsPSY2-EYFP signal were detected using the anti-GFP antibodies, both these two proteins located in the thylakoid membrane fraction but not stroma and envelope fractions compared to marker proteins (Tic110, envelope; RbcL, stroma; Lhca1, thylakoid membrane). Env, envelope membranes; Str, stroma; Tm, thylakoid membrane.
Further immunoblot analysis using different sub-fractions prepared from Percoll-purified chloroplasts extracted from above infiltrated leaves revealed presence of both OsLIL3-EYFP and OsPSY2-EYFP in the thylakoid membranes, but not in the stromal fraction (Fig. 5b).
OsLIL3 Induces Gene Expression of its Interaction Partners Involved in Carotenoid and Chlorophyll Biosynthesis
To further analyze the gene expression of enzymes involved in carotenoid and chlorophyll biosynthesis, 15 candidate genes were selected from DEGs in these pathways. qRT-PCR validation of these genes indicated that gene expression of OsLIL3-interacting genes such as OsPSY (OsPSY1, OsPSY2, and OsPSY3), OsGGR, OsPORA, OsOR, OsCLP (OsCLPC1&OsCLPS1) family genes was greatly suppressed (Fig. 6).
Expression analysis of genes in carotenoid biosynthesis and validation of genes identified in RNA-seq analysis. qRT-PCR analysis of genes related to carotenoid biosynthesis, chlorophyll biosynthesis and validation of genes identified as DEGs in oslil3 vs WT this transcriptome analysis. Two reference genes were used as internal controls. The expression levels of genes in WT plants were set to 1. OsAtpB, Os10g21266; OsPSY1, Os06g51290; OsPSY2, Os12g43130; OsPSY3, Os09g38320; OsGGR, Os02g51080; OsPORA, Os04g58200; OsPORB, Os10g35370; OsNECD, Os12g24800; OsLUT1, Os10g39930; OsCPOX, Os04g52130; OsCHLI, Os03g36540; OsOR, Os02g43500; OsCLPC1, Os04g32560; OsCLPS1, Os08g33540; OsLHCA2, Os09g26810
Discussion
Carotenoids refer to a large number of yellow, orange or lipophilic molecules that are synthesized during photosynthesis (Rodriguez-Concepcion et al. 2018). The various functional roles of carotenoids involve: light harvest/capture; protection against abiotic stress conditions (Dall’Osto et al. 2007; Stanley and Yuan 2019); acting as a precursor in hormone biosynthesis (such as for biosynthesis of GA that acts as a stress and developmental signal) (Walter et al. 2010; Sun et al. 2018), quenching of singlet oxygen in photosynthesis II under normal conditions (Tamura and Ishikita 2020), and rendering tolerance to heat and light stress by protecting membranes from reactive oxygen species (ROS) and lipid peroxidation (Johnson et al. 2007; Yokono and Akimoto 2018).
In our study, the CRISPR-Cas9 oslil3 mutant was used for a detailed physiological, transcriptomic, and molecular characterization of rice OsLIL3 gene. The mutant has a 4-bp deletion leading to a frameshift mutation. It displayed a yellowish leaf color phenotype that is more prominent in the seedling stages (Fig. 1a, b and c). oslil3 contained reduced levels of chlorophylls and carotenoids in the mutant vs WT. In addition, OsLIL3 mutation caused aberrant chloroplast development under normal growth conditions. In Arabidopsis, the lil3.1/lil3.2 double mutant showed reduced chlorophyll contents while the single lil3.1 or lil3.2 mutants displayed no significant differences from WT (Hey et al. 2017). Since OsLIL3 mutation leads to a strong reduction in chlorophyll and carotenoid levels it could act as a master regulator of any downstream stress caused by a reduction in these pigment levels. Previous investigation of LIL3 homologs in dicot (Arabidopsis AtLIL3.1 and AtLIL3.2) and monocot (rice OsLIL3) revealed a conserved function in chlorophyll and terpenoid biosynthesis through their interaction with GGR and POR enzymes (Tanaka et al. 2010; Hey et al. 2017; Zhou et al. 2017). LIL3 proteins interact directly with different isoforms of POR and GGR enzymes and regulate their protein abundances in rice and Arabidopsis (Hey et al. 2017; Tanaka et al. 2010). The yellow leaf phenotype of lil3.1/lil3.2 mutant may be caused by the reduced protein levels of POR and GGR enzymes involved in chlorophyll and terpenoid biosynthesis. POR is responsible for reduction of protochlorophyllide to chlorophyllide and GGR is responsible for the generation of intermediate metabolite GGPP (Tanaka et al. 2010; Hey et al. 2017). A similar case may be made for the yellow leaf phenotype of rice oslil3. However, the molecular relation between LIL3 levels and these pathway proteins is still disconnected.
A genome wide transcriptome analysis of oslil3 identified that of 4288 DEGs in total, 2188 genes were up-regulated and 2100 genes were down-regulated vs WT (Fig. 2a). GO enrichment analysis suggested that OsLIL3 deficiency leads to expression changes in genes localized to thylakoid membrane, lumen and PSII; in genes involved in aldonate transmembrane transport, chitin metabolism and many amino-sugar catabolic pathways and for genes with chitinase, transporter and sugar modification activity. KEGG pathway analysis suggested similarly that many metabolic pathways are affected in the oslil3 mutant especially plant-pathogen interaction genes (possibly chitin related genes) and in carbon metabolism, photosynthetic light reactions, and isoprenoid pathways and so on (Figure S3 and S4). Interestingly, all of the DEGs mapped in the MapMan reveals that genes in the cell wall biosynthesis were down-regulated and most of the genes in light reactions were up-regulated (Fig. 3a &b). Interestingly, four CESA proteins (CESA1, CESA3, CESA4, CESA6) that are the core components of the cell wall cellulose synthase (Kumar et al. 2016) were down-regulated that will repress the synthetic activities of cellulose in oslil3. COBL4 is also another reported gene responsible for cellulose biosynthesis in primary and secondary cell wall and help crystallize cellulose in the walls (Brown et al. 2005). Six FLA genes were also identified as down-regulated in oslil3, however, the functional roles of FLA family proteins are largely unknown. The FLA3 were reported to be involved in cellulose biosynthesis in pollen tubes (Andersson-Gunnerås et al. 2006). This could explain the dwarf phenotype observed in oslil3. There were distinct differences between chloroplasts from oslil3 and WT based on ultrastructural and physiological characteristics. RNA-seq data comparison of the wp1 (Wang et al. 2016) and oslil3 mutant revealed a common pattern of downregulation of secondary metabolic process (phenylpropanoid biosynthesis, flavonoid biosynthesis), lignin biosynthetic process, steroid biosynthetic process, and of up-regulation of chlorophyll organization, amino sugar and nucleotide sugar metabolism and chitin and polysaccharide catabolism. We can conclude that both LIL3 and WP1 are involved in chloroplast organization and pigment biosynthesis that in turn play essential roles in plant growth and in determining the leaf color.
In Arabidopsis, previously it was reported that deficiency of LIL3 levels leads to reduced accumulation level of GGR protein and influences chlorophyll biosynthesis that seemed to support a similar yellowish phenotype. Arabidopsis LIL3 mutation down-regulated genes involved in light reaction pathway and isoprenoid pathways under light stress conditions similar to the findings from our oslil3 transcriptomic data (Lohscheider et al. 2015). It also showed reduced abundance of POR enzymes similar to our immunoblot analysis findings (Hey et al. 2017). This suggests a common mechanism of lower abundances of GGR or POR enzymes leading to the yellow leaf phenotype in both studies. Previous studies suggest how rice LIL3 directly regulates isoprenoid enzymes (leading to a yellow leaf phenotype). In rice, LIL3 interacts with POR, GGR, geranylgeranyl diphosphate synthase (GGDPPS1) and GGPPS recruiting protein (GRP) proteins that are components of a large multiprotein complex in the thylakoid membrane (Zhou et al. 2017). Looking for additional LIL3-novel interacting proteins in the isoprenoid biosynthesis pathway revealed that OsPSY2, phytoene synthase 2, shows positive interaction with OsLIL3 in this study (Fig. 4). The other two ortholog genes namely OsPSY1 and OsPSY3, also have positive interactions with OsLIL3. Phytoene synthase is a major rate limiting enzyme in carotenoid biosynthesis pathway that is controlled by the Clp protease family proteins and ORGANLE (OR) protein in the plastid carotenoid biosynthesis in Arabidopsis (Welsch et al. 2018; Zhou et al. 2015b). The OR protein is a post-transcriptional regulator of PSY while the Clp protease degrades and controls the PSY protein turnover in Arabidopsis. LIL3 contains typical LHC domain characterized by about four hydrophobic and α-helical membrane-spanning domains that can bind dozens of chlorophyll and three to four different carotenoids (Zapata et al. 2000), thus playing vital roles in photosynthesis and photoprotection. Alteration of PSY expression displayed profound effects on the content of carotenoids. A number of factors were reported to have influences on the expression of PSY gene expression (Cheminant et al. 2011; Toledo-Ortiz et al. 2010). For example, the expression of PSY1 in tomato was regulated by carotenoids and transcriptional factors like RIPENING INHIBITOR (Martel et al. 2011). Gibberellin-regulated DELLA protein regulated the expression of genes involved in carotenoid biosynthesis during de-etiolation process in Arabidopsis (Cheminant et al. 2011). The tomato ERF gene, SIAP2α, regulated the carotenoid accumulation profile by altering carotenoid pathway flux (Chung et al. 2010). Recently, PSY protein levels were shown to be regulated by a negative feedback emerging from carotenoids (Arango et al. 2014). However, the factors that may act as translational or post-transcriptional regulators are still limited. In our study, the mRNA levels of OsPSY1, OsPSY2, and OsPSY3 was significantly down-regulated in oslil3 mutant as compared to WT (Fig. 6) and a positive interaction between LIL3 and PSY proteins which suggests that LIL3 may act as a post-transcriptional regulator of PSYs, representing an important regulatory mechanism underlying carotenoid biosynthesis in plants. There is a possibility that OsPSY genes may be another example of LIL3 anchored protein partner in the chloroplast similar to GGR and POR (Tanaka et al. 2010; Hey et al. 2017). Indeed, the evidence from subcellular localization analysis suggests OsPSY2 and OsLIL3 are both co-localized to thylakoid membranes in the chloroplasts (Fig. 5). No changes were found in the expression levels of OR while the CLPC1 and CLPS1 were significantly down-regulated in oslil3 mutant. The expression levels of PSY interacting genes (OR, CLPC1 and CLPS1) varied and showed different pattern in oslil3 mutant while PSY genes were all down-regulated, suggesting that LIL3 may also act as a regulator of PSY mRNA in addition to interacting with it.
Interestingly, reduced protein contents of POR as well as gene expression of other genes in chlorophyll and carotenoid do not correlate well with the levels of the transcription of their corresponding genes and may be due to post-translational modifications (Hey et al. 2017). However, there is a possibility that OsLIL3 may act as regulator for pigments genes other than POR and GGR enzyme genes in plants (Tanaka et al. 2010; Hey et al. 2017; Zhou et al. 2017). Indeed, we found in our immunoblot study that OsLIL3 enhanced the protein levels of OsPORB & OsGGR, that is highly consistent with the previous findings (Tanaka et al. 2010; Hey et al. 2017). The repressed protein abundances of PORB and GGR enzymes lead to lower levels of chlorophyll contents and subsequently caused yellow leaf phenotype in several studies (Kimura et al. 2018; Li et al. 2019; Buhr et al. 2017; Liu et al. 2015). On the other hand, overexpression of PORB produces more chlorophyll metabolites in plant leaves (Zhan et al. 2019) and overexpression of GGR also generates more contents of chlorophylls in tomato leaves (Liu et al. 2015). The chlorophyll contents are reduced in oslil3 mutant that is in accordance with the lower accumulation levels of PORB and GGR protein in this mutant.
Meanwhile, qPCR (Figure S8) also revealed that the relative mRNA expression levels of OsPORA, OsGGR, OsOR and OsCLP are greatly suppressed in oslil3 mutant vs WT while the OsPORB expression was about 50% lower (Fig. 6). This differential expression pattern of OsPORA was also detected in our RNA-seq analysis but expression level OsPORB did not significantly change in WT and oslil3. As reported, the OsPORA mainly functions in the early stages of leaf development while OsPORB is expressed consistently throughout and is essential for maintaining light-dependent chlorophyll biosynthesis throughout leaf development (Sakuraba et al. 2013). This is in argument with the observation that oslil3 displays a prominent yellow leaf phenotype only in the early seedling stages but not in adult stages. In Arabidopsis, the protein accumulation level of PORA and PORB was greatly suppressed in lil3.1/lil3.2 double mutant while PORC was depressed to a lesser extent. Interestingly, expression of OsOR did not significantly change in our RNA-seq and qRT-PCR experiment (Fig. 6). However, OsOR protein was reported to be a key post-transcriptional regulator of PSY in carotenoid biosynthesis (Zhou et al. 2015a). This suggests that the regulation of PSY levels may be determined by multiple molecular regulators such as OR and LIL3. Further studies need to investigate the protein accumulation levels of other PSY interacting proteins. Protein-protein interaction assays are required to determine whether the interaction of LIL3-PSY is uniquely essential in controlling carotenoid biosynthesis. Additionally, the regulation of enzymatic activities of PSYs in oslil3 mutant and protein levels of LIL3 in Ospsy mutants are also necessary to be determined to further support our findings.
As we have drafted the abbreviated model of chlorophyll, carotenoid and GA biosynthesis in Fig. 3b, these three pathways are connected by GGPP (Geranylgeranyl diphosphate, an intermediate in these pathway). GGPP acts as an entry point to the biosynthesis of carotenoids, chlorophylls, tocopherols, gibberellins, plastoquinones, polyprenols, diterpenoids, and mitochondria-derived polyterpenes (Nagegowda and Gupta 2020). Overexpression of PORA, PORB or PORC could accumulate more Chls and protect plants against photooxidative damage in Arabidopsis (Pattanayak and Tripathy 2011; Sperling et al. 1997). In crop plants, overexpression of PORB also improves chlorophyll and tocopherol content in both leaves and kernels in maize (Zhan et al. 2019). Overexpression of PSY induced accumulation of phytoene but not of any downstream products in rice (Burkhardt et al. 1997). Thus, any metabolic engineering of the carotenoid biosynthesis could also influence the chlorophylls but would still play positive roles in plant growth and performance under oxidative stress conditions in plants. In summary, our work revealed a novel functional impact of OsLIL3 mutation on carotenoid and chlorophyll biosynthesis. OsLIL3 may regulate carotenoid and chlorophyll biosynthesis not only through its interaction with a thylakoid co-localizing OsPSY2 but also by enhancing the protein levels (OsPORB, OsGGR) and regulating the gene expression (OsPSYs, OsGGR, OsPORA, OsLUT1, OsCPOX, OsCHLI and OsCLP) of enzymes involved in these pathways. Thus, LIL3 may serve as a common hub for modulating levels of enzymes involved in different plant pigment pathways to manipulate its metabolism and growth.
Materials and Methods
Plant Materials and Growth Conditions
The CRISPR-Cas9 mutant oslil3 (Oryza sativa ssp. japonica cv. Nipponbare) was generated using CRISPR-Cas9 system at Biogle (http://www.biogle.cn/, Hangzhou, China). A single sgRNA targeting OsLIL3 at the location 5′-GGCGCCTCGACAGTCTCCACGGG-3′ was created in the BGK03 vector (Bai et al. 2020) containing Cas9, that was then transformed into Agrobacterium tumefaciens strain EHA105 and the transgenic rice plants were obtained as mentioned previously (Nishimura et al. 2006). Genomic DNA from these transformants was sequenced for verifying precise mutation site in the OsLIL3 sequence using PCR amplification with primer pairs flanking the target site and potential off-targets were also investigated based on the online prediction tool CRISPR-P (http://cbi.hzau.edu.cn/cripr/) (Supplementary data S1). The mutant line 16-#1, (T2 stable transgenic CRISPR-Cas9 plant), is a 4-bp deletion OsLIL3 knock-out mutant as compared to the other lines. As analyzed in the segregation experiment, the 3-bp and 9-bp deletions did not cause any frameshift in translation while only the 4-bp deletion caused a frameshift premature translation termination (Supplementary data S2). This mutant has a chlorotic phenotype and was used for further analysis. Amplified PCR products of size (300–500 bp) were sequenced directly and analyzed using the Degenerate Sequence Decoding method (Ma et al. 2015) to identify the mutated sites.
TEM Analysis of Chloroplast Structures
Third leaf from 1-week-old WT and oslil3 mutant seedlings (3-leaf stage) grown at 28 °C were used to prepare samples for transmission electron microscopy (TEM, Philips CM100, Eindhoven, The Netherlands) analysis. Leaf samples were fixed in 2.5% glutaraldehyde cacodylate buffer at 4 °C for 12 h. The fixation was stopped by changing to cacodylate buffer containing 0.1 M sucrose. All subsequent experiments were carried out in the fume hood. Leaf samples were fixed in 1% osmium tetroxide cacodylate buffer at 28 °C for 2 days, and samples were washed several times with cacodylate buffer (0.05 M sodium cacodylate in an aqueous solution at pH 7.2–7.4). A dehydration procedure was performed as follows: a) 50% ethanol (10 min), 70% ethanol (10 min), 90% ethanol (10 min), 100% ethanol (20 min) (step repeated 3 times); b) 99% propylene oxide (10 min) (step repeated 2 times). The tissues were then infiltrated with 1:2 mixture of epoxy resin/propylene oxide for 1.5 h, 1:1 mixture of epoxy resin/propylene oxide for 1.5 h. and 2:1 mixture of epoxy resin/propylene oxide overnight at 28 °C. Then, the samples were infiltrated with fresh epoxy resin for 3 h at 37 °C in a vacuum oven. These samples were finally embedded in fresh epoxy resin in plastic capsules and polymerized at 60 °C overnight. Ultra-thin sections were visualized under the transmission electron microscope (Philips CM100, Eindhoven, The Netherlands).
RNA Extraction and Quality Control
WT and oslil3 mutant plant samples were collected at the L3 seedling stage and whole leaf samples were harvested for RNA extraction (3 biological replicates with 6–8 individual plants for each replicate). Leaves from oslil3 and WT were rapidly frozen in liquid nitrogen and total RNA was extracted with the EZNA plant RNA kit (Omega Bio-Tek, GA, USA) according to the manufacturer’s instructions. RNA quantity and purity were checked by the kaiaoK5500 Spectrophotometer (Kaiao, Beijing, China). RNA integrity and concentration were analyzed with the RNA Nano 6000 Assay Kit of Bioanalyzer 2100 (Agilent Technologies, CA, USA). RNA concentration for library construction was measured using Qubit RNA Assay Kit in Qubit 3.0 (Life Technologies, Grand Island, NY, USA). Dilutions were prepared at a concentration of 1 μg·μl− 1.
Library Preparation and Sequencing
For library preparation, 2 μg total RNA was input into NEB Next Ultra RNA library Prep Kit (New England Biolabs, NEB, USA) to generate sequencing libraries based on the manufacturer’s recommendation. In brief, mRNA was purified from the input total RNA using poly-dT oligo-attached magnetic beads. This was followed by RNA fragmentation by addition of divalent cations under increased temperature. First strand cDNA synthesis (Takara, Japan) was performed on the fragmented RNA using random hexamer primers and the remaining RNA was degraded using RNase H (Omega Bio-Tek, USA). Subsequently, a second strand cDNA synthesis was performed and the resulting fragments were purified using the QiaQuick PCR kit (QIAgen, Hiden, Germany) followed by terminal repair, A-tailing and adapter addition. Finally, PCR reactions were carried out to complete library preparation.
Library insert size was quantified using StepOnePlus Real-Time PCR system (library valid concentration > 10 nmol·L-1). Sample clustering was performed on a cBot cluster generation system using HiSeq PE Clustering Kit v4-cBot-HS (Illumina, CA, USA). Subsequently, libraries were sequenced on a Illumina HiSeq 4000 platform (Illumina, CA, USA) to obtain 150 bp paired-end reads.
Transcriptome Analysis and Differential Gene Expression
Reads were processed through quality check using FastQC protocol and high-quality reads (“clean reads”) were obtained for mapping to reference genome (ftp://ftp.ensemblgenomes.org/pub/plants/release-24/fasta/oryza_sativa/) in HISAT2 v2.0.5 (Kim et al. 2015). Following this, raw read count for each gene in each sample was obtained with HTSeq v0.6.0, and FPKM (Fragments Per Kilobase Million mapped reads) was calculated from the raw read counts to estimate the gene expression level of all the expressed genes in each sample.
Principal component analysis (PCA) analyses were conducted to assess the transcriptome variance in sample replicates (Tsuyuzaki et al. 2020). Hierarchical clustering organizes samples (experimental units) based on the gene expression within each sample using Poisson model and dissimilarity measure based on likelihood ratio statistics (Si et al. 2014).
Finally, DESeq2 v1.6.3 was used to compare gene expression levels between oslil3 and WT. Gene expression level was determined by linear regression calculating the fold changes for sample comparisons. Subsequently, p-value for this fold change was calculated with Wald test. Finally, these p-values were corrected by the BH method to give q-values. Genes passing both cut-offs of q-value ≤0.05 and |log2 fold change| ≥ 1 were identified as differentially expressed genes (DEGs). Functional enrichment analyses were performed using MapMan analysis (https://mapman.gabipd.org/), GO functional classification (http://www.blast2go.org/) and KEGG pathway analyses (https://www.genome.jp/kegg/) as performed previously (Song et al. 2020). MapMan analysis maps the DEGs according to the log fold change values on the pathway maps. The rich-ratio in GO analysis is defined as amount of differentially expressed genes enriched in the pathway/amount of all genes in that pathway in the background gene set. The size of the dots represents the number of genes, and the color of the dots represents the false discovery rate (FDR) (Zhang et al. 2020). KEGG analyses plots the number of genes belonging to enriched pathways.
Y2H Assay
Coding regions of OsLIL3 and OsPSY genes were cloned without transit peptide and transmembrane domains and fused in-frame into the bait vector pGBKT7 or prey vector pGADKT7 with corresponding primers (Clontech, Mountain View, USA) (Supplementary data S3) and further verified for correct sequence by DNA sequencing. Yeast transformation and selection procedures were performed according to the protocols of the Matchmaker gold yeast two-hybrid (Y2H) system (Clontech, Mountain View, USA). S. cerevisiae Gold cells (Clontech, Mountain View, USA) that were transformed with different combinations of bait and prey vector constructs were incubated on synthetic defined (SD) plates lacking Leu and Trp for selection of co-transformants. Afterwards, the co-transformed yeast cells were further selected on high selection plates (SD –Leu –Trp –His + X-α-Gal + Aureobasidin A) to identify blue colonies indicative of positive protein-protein interactions.
BiFC Assay
BiFC is commonly used for detection of protein-protein interaction in living mammalian tissue culture and the fluorescent protein YFP and other GFP derivatives are split into non-overlapping N-terminal or C-terminal part. Full-length coding regions of OsLIL3 were cloned in-frame with the N-terminal fragment of EYFP in the SPYNE vector (amino acids 1–155, pSPYNE). On the other hand, full-length coding regions of different candidate proteins OsPSYs were cloned in-frame with the C-terminal fragment of EYFP in the SPYCE (amino acids 156–239, pSPYCE) binary vector. Both construct sequences were confirmed with DNA sequencing. Different combinations of BiFC vectors were transiently expressed in N. benthamiana leaves by Agrobacterium infiltration as described previously (Sparkes et al. 2006). Fluorescent signals in the infiltrated leaves were examined after 48–72 h with a confocal microscope (LSM710, Carl Zeiss Microscopy, Germany).
Co-Immunoprecipitation Assay
Co-immunoprecipitation assay was performed according to the methods described in Jin et al. 2018 with some minor modifications. Briefly, the constructs containing the full length of OsLIL3-FLAG and OsPSYs-HA were generated with PCR amplification and correct gene sequence was confirmed by DNA sequencing. In addition, the combinations of co-infiltration were OsLIL3:FLAG and OsPSY1:HA, OsLIL3:FLAG and OsPSY2-HA, or OsLIL3:FLAG and OsPSY3:HA. These construct pairs were transformed into N. benthamiana leaves with agro-infiltration (agrobacterium suspensions containing the respective constructs is infiltrated) and after 48 h incubation, the total proteins were extracted from infected leaf areas and solubilized in lysis buffer [10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 0.5% NP-40] with 1 mM protease inhibitor cocktail (Sigma-Aldrich, USA) and 1 mM PMSF (Sigma-Aldrich, USA). The solubilized total proteins were incubated with anti-HA agarose beads (Thermo Scientific, USA) for 1 h at 4 °C. The supernatant was discarded after centrifugation at 2500 rpm for 2 min at 4 °C. The remaining pellets were rinsed with washing buffer [10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.25% NP-40] for at least three times and the beads were transferred to a fresh tube, and subsequently resuspended in a 100 μl 2x SDS-sample buffer [100 mM Tris HCl, pH 6.8, 200 mM dithiothreitol, 4% SDS, 0.2% bromophenol blue, 20% glycerol] and boiled. The supernatant was collected for immunoblotting with monoclonal anti-FLAG (1:10000; Sigma-Aldrich, USA), and monoclonal anti-HA anti-body (1:5000, Abcam, Cambridge, UK). Signals were detected using an ECL Western Blotting Detection Kit (Amersham Life Science, Little Chalfont, Buchinghamshire, England) on Kodak X-ray films.
Immunoblot Analyses of OsLIL3, OsGGR, OsPORB in oslil3
WT and oslil3 mutant rice samples were prepared at the L3 seedling stage and used for immunoblot analysis. To isolate total proteins, rice leaves were ground in ice cold isolation buffer (50 mM Tris–HCl pH 7.8, 250 mM sucrose, 25 mM KCl, 10 mM MgCl2, 0.5% (v/v) β-mercaptoethanol, 0.5% (v/v) Triton X-100) and were centrifuged at 13000 g for 10 min. Supernatants were collected and used to extract protein samples for immunoblot experiments. Protein extracts (30 μg) were subjected to SDS-PAGE and immunoblotting as described (Lee et al., 2013). Anti-OsLIL3 antibodies (Kang Ti, Shenzhen, China) were generated in the rabbits against an oligopeptide, AMIGFFMAYFVDSL, that corresponds to amino acid residues of OsLIL3 at positions 172 to 185. Immunoblotting was performed using rabbit polyclonal antibodies against OsLIL3 (1:2000), GGR (1:10000, Agrisera, Vännäs, Sweden), POR (1:5000; Agrisera) or AtpB (1:30000;). The membranes were incubated with horseradish peroxidase–conjugated goat anti-rabbit antibodies (1:25,000; Agrisera, Vännäs, Sweden). Signals were detected using an ECL Western Blotting Detection Kit (Amersham, Little Chalfont, Buchinghamshire, England) on Kodak X-ray films.
Subcellular Localization of OsLIL3 and OsPSY2
The OsLIL3 and OsPSY2 coding region were cloned in-frame with the EYFP gene in the pCAMBIA1301-35S-NOS vectors as previously described (Li et al. 2014). Agrobacterium tumefaciens (strain GV3101) cultures harboring either 35S::OsLIL3-EYFP and 35S::OsPSY2-EYFP constructs were adjusted to OD600 = 0.8 in MES buffer (10 mM MES, pH 5.5, and 10 mM MgSO4) and infiltrated into leaves of N. benthamiana (6-week-old). For agroinfiltration in N. benthamiana, A. tumefaciens (strain GV3101) cultures harboring constructs were incubated in 28 °C in 10 ml LB with 100 μg·ml− 1 kanamycin for 2 days. Cells were pelleted at 4000 rpm for 15 min, and re-suspended in induction medium at 28 °C overnight. Fluorescent signal in the infiltrated leaves was examined after 3 days by confocal microscopy (LSM710 Carl Zeiss Microscopy, Germany).
Chloroplasts were then isolated from infiltrated N. benthamiana leaves. Stroma, envelope, and thylakoid membrane fractions were prepared as described previously (Salvi et al. 2008). Protein samples from intact chloroplast as well as the stroma, envelope and thylakoid fractions were separated by 10% SDS-PAGE gel for electrophoretic analysis. The proteins were then transferred onto nitrocellulose membranes (Amersham, Little Chalfont, Buchinghamshire, England). Immunoblot experiments were performed with polyclonal mouse antibodies against GFP (1:3000 dilution; Invitrogen, Life technologies, Carls bad, CA, USA) and rabbit polyclonal antibodies against RbcL, Lhca1, Tic110 (1:5000, 1:2000, 1:1000 dilution, respectively; Agrisera, Vännäs, Sweden), followed by incubation with horseradish peroxidase–conjugated goat anti-mouse IgG antibodies (1:3000 dilution; Invitrogen, Life technologies, Carls bad, CA, USA), or goat anti-rabbit antibodies (1:25,000; Agrisera, Vännäs, Sweden).
qRT-PCR Validation Assays
Validation of RNA-seq results was performed for 15 genes selected from DEGs that are involved in photosynthesis and other pathways using quantitative RT-PCR (qRT-PCR). The sequences of the gene primers are provided in Supplementary data S3. Total RNA was extracted as mentioned above for RNA-Seq and first-strand cDNA was synthesized using a TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit utilizing oligodT primers (TransGen Biotech, Beijing, China). qRT-PCR was carried out in the ABI Step One Plus Real-Time PCR System (Applied Biosystems, USA) utilizing the SYBR Premix Ex Taq RT-PCR kit (Takara Bio, Japan). qRT-PCR programs were as follows: (i) 95 °C for 30 s, (ii) 40 cycles of denaturation at 95 °C for 5 s, annealing at 60 °C for 10s and extension at 72 °C for 10s. Rice ATP synthase subunit beta (OsAtpB, Os10g21266) gene was employed as negative controls in this study. The two rice reference gene Osactin1 (Os03g50885) and Osubiquitin (Os03g03920) were used as internal control genes for expression normalization.
Statistical Analysis
Statistics was conducted using IBM SPSS Statistics ver. 18 (SPSS Inc., IL, USA) employing Duncan’s Multiple Range test (DMRT) post-hoc test with significance level set to P ≤ 0.05 for sample comparisons. Data is mean ± SE from three independent biological replicates. Student’s t-test was performed to employ the data analysis of the qRT-PCR assay.
Availability of Data and Materials
All relevant data are provided within the article and its supplementary information files.
Change history
12 May 2021
A Correction to this paper has been published: https://doi.org/10.1186/s12284-021-00484-x
Abbreviations
- LIL3:
-
Light harvesting like protein 3
- GGR:
-
Geranylgeranyl reductase
- POR:
-
Protochlorophyllide oxidoreductase
- PSY:
-
Phytoene synthase
- FC:
-
Ferrochelatase
- GGPPS1:
-
Geranylgeranyl diphosphate synthase
- CHLG:
-
Chlorophyll synthase
- RNA-seq:
-
RNA-sequencing
- Y2H:
-
Yeast two hybrid assay
- BiFC:
-
Bimolecular fluorescence complementation
- CoIP:
-
Co-immunoprecipitation assay
- TEM:
-
Transmission electron microscopy
- PS I:
-
photosystem I
- PS II:
-
Photosystem II
- LHC:
-
Light harvesting complex
References
Adhikari ND, Froehlich JE, Strand DD, Buck SM, Kramer DM, Larkin RM (2011) GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23(4):1449–1467. https://doi.org/10.1105/tpc.110.082503
Andersson-Gunnerås S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45(2):144–165. https://doi.org/10.1111/j.1365-313X.2005.02584.x
Arango J, Jourdan M, Geoffriau E, Beyer P, Welsch R (2014) Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in orange carrots. Plant Cell 26(5):2223–2233. https://doi.org/10.1105/tpc.113.122127
Bai M, Yuan J, Kuang H, Gong P, Li S, Zhang Z, Liu B, Sun J, Yang M, Yang L, Wang D, Song S, Guan Y (2020) Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol J 18(3):721–731. https://doi.org/10.1111/pbi.13239
Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17(8):2281–2295. https://doi.org/10.1105/tpc.105.031542
Buhr F, Lahroussi A, Springer A, Rustgi S, von Wettstein D, Reinbothe C, Reinbothe S (2017) NADPH: protochlorophyllide oxidoreductase B (PORB)action in Arabidopsis thaliana revisited through transgenic expression of engineered barley PORB mutant proteins. Plant Mol Biol 94(1):45–59
Burkhardt PK, Beyer P, Wünn J, Klöti A, Armstrong GA, Schledz M, von Lintig J, Potrykus I (1997) Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin a biosynthesis. Plant J 11(5):1071–1078. https://doi.org/10.1046/j.1365-313X.1997.11051071.x
Cheminant S, Wild M, Bouvier F, Pelletier S, Renou J-P, Erhardt M, Hayes S, Terry MJ, Genschik P, Achard P (2011) DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23(5):1849–1860. https://doi.org/10.1105/tpc.111.085233
Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64(6):936–947. https://doi.org/10.1111/j.1365-313X.2010.04384.x
Dall'Osto L, Cazzaniga S, North H, Marion-Poll A, Bassi R (2007) The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. Plant Cell 19(3):1048–1064. https://doi.org/10.1105/tpc.106.049114
Deng X-j, H-q Z, Wang Y, He F, Liu J-L, Xiao X, Shu Z-F, Li W, Wang G-H, Wang G-L (2014) Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PloS One 9(6):e99564
Funk C, Vermaas W (1999) A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38(29):9397–9404. https://doi.org/10.1021/bi990545+
Hey D, Rothbart M, Herbst J, Wang P, Müller J, Wittmann D, Gruhl K, Grimm B (2017) LIL3, a light-harvesting complex protein, links terpenoid and tetrapyrrole biosynthesis in Arabidopsis thaliana. Plant Physiol 174(2):1037–1050. https://doi.org/10.1104/pp.17.00505
Jansson S (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4(6):236–240. https://doi.org/10.1016/S1360-1385(99)01419-3
Johnson MP, Havaux M, Triantaphylidès C, Ksas B, Pascal AA, Robert B, Davison PA, Ruban AV, Horton P (2007) Elevated zeaxanthin bound to oligomeric LHCII enhances the resistance of Arabidopsis to photooxidative stress by a lipid-protective, antioxidant mechanism. J Biol Chem 282(31):22605–22618. https://doi.org/10.1074/jbc.M702831200
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. https://doi.org/10.1038/nmeth.3317
Kimura E, Abe T, Murata K, Kimura T, Otoki Y, Yoshida T, Miyazawa T, Nakagawa K (2018) Identification of OsGGR2, a second geranylgeranyl reductase involved in α-tocopherol synthesis in rice. Scientific reports 8(1):1–8
Kumar M, Campbell L, Turner S (2016) Secondary cell walls: biosynthesis and manipulation. J Exp Bot 67(2):515–531. https://doi.org/10.1093/jxb/erv533
Li C, Liu X, Pan J, Guo J, Wang Q, Chen C, Li N, Zhang K, Yang B, Sun C (2019) A lil3 chlp double mutant with exclusive accumulation of geranylgeranyl chlorophyll displays a lethal phenotype in rice. BMC Plant Biol 19(1):456. https://doi.org/10.1186/s12870-019-2028-z
Li Q, Zhu F-Y, Gao X, Sun Y, Li S, Tao Y, Lo C, Liu H (2014) Young leaf Chlorosis 2 encodes the stroma-localized heme oxygenase 2 which is required for normal tetrapyrrole biosynthesis in rice. Planta 240(4):701–712. https://doi.org/10.1007/s00425-014-2116-0
Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen L-L (2017) CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10(3):530–532. https://doi.org/10.1016/j.molp.2017.01.003
Liu H, Liu J, Zhao M-M, Chen J-S (2015) Overexpression of ShCHL P in tomato improves seedling growth and increases tolerance to salt, osmotic, andoxidative stresses. Plant Growth Regul 77(2):211–221
Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W (2004) Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428(6980):287–292. https://doi.org/10.1038/nature02373
Lohscheider JN, Rojas-Stütz MC, Rothbart M, Andersson U, Funck D, Mendgen K, Grimm B, Adamska I (2015) Altered levels of LIL 3 isoforms in Arabidopsis lead to disturbed pigment–protein assembly and chlorophyll synthesis, chlorotic phenotype and impaired photosynthetic performance. Plant Cell Environ 38(10):2115–2127. https://doi.org/10.1111/pce.12540
Ma X, Chen L, Zhu Q, Chen Y, Liu Y-G (2015) Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol Plant 8(8):1285–1287. https://doi.org/10.1016/j.molp.2015.02.012
Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157(3):1568–1579. https://doi.org/10.1104/pp.111.181107
Mork-Jansson AE, Eichacker LA (2018) Characterization of chlorophyll binding to LIL3. PLoS One 13(2):e0192228. https://doi.org/10.1371/journal.pone.0192228
Nagata N, Tanaka R, Satoh S, Tanaka A (2005) Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17(1):233–240. https://doi.org/10.1105/tpc.104.027276
Nishimura A, Aichi I, Matsuoka M (2006) A protocol for agrobacterium-mediated transformation in rice. Nat Protoc 1(6):2796–2802. https://doi.org/10.1038/nprot.2006.469
Pattanayak GK, Tripathy BC (2011) Overexpression of protochlorophyllide oxidoreductase C regulates oxidative stress in Arabidopsis. PLoS One 6(10):e26532. https://doi.org/10.1371/journal.pone.0026532
Rodriguez-Concepcion M, Avalos J, Bonet ML, Boronat A, Gomez-Gomez L, Hornero-Mendez D, Limon MC, Meléndez-Martínez AJ, Olmedilla-Alonso B, Palou A (2018) A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res 70:62–93. https://doi.org/10.1016/j.plipres.2018.04.004
Sakuraba Y, Rahman ML, Cho SH, Kim YS, Koh HJ, Yoo SC, Paek NC (2013) The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J 74(1):122–133. https://doi.org/10.1111/tpj.12110
Salvi D, Rolland N, Joyard J, Ferro M (2008) Purification and proteomic analysis of chloroplasts and their sub-organellar compartments. In: Organelle Proteomics. Springer, pp 19–36
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9(10):2395–2410. https://doi.org/10.1038/nprot.2014.157
Shao C-H, Qiu C-F, Qian Y-F, Liu G-R (2020) Nitrate deficiency decreased photosynthesis and oxidation-reduction processes, but increased cellular transport, lignin biosynthesis and flavonoid metabolism revealed by RNA-Seq in Oryza sativa leaves. PLoS One 15(7):e0235975. https://doi.org/10.1371/journal.pone.0235975
Shim K-C, Kim SH, Jeon Y-A, Lee H-S, Adeva C, Kang J-W, Kim H-J, Tai TH, Ahn S-N (2020) A RING-type E3 ubiquitin ligase, OsGW2, controls chlorophyll content and dark-induced senescence in Rice. Int J Mol Sci 21(5):1704. https://doi.org/10.3390/ijms21051704
Si Y, Liu P, Li P, Brutnell TP (2014) Model-based clustering for RNA-seq data. Bioinformatics 30(2):197–205. https://doi.org/10.1093/bioinformatics/btt632
Song T, Das D, Yang F, Chen M, Tian Y, Cheng C, Sun C, Xu W, Zhang J (2020) Genome-wide transcriptome analysis of roots in two rice varieties in response to alternate wetting and drying irrigation. Crop J 8(4):586–601. https://doi.org/10.1016/j.cj.2020.01.007
Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1(4):2019–2025. https://doi.org/10.1038/nprot.2006.286
Sperling U, Van Cleve B, Frick G, Apel K, Armstrong GA (1997) Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J 12(3):649–658. https://doi.org/10.1046/j.1365-313X.1997.d01-11.x
Stanley L, Yuan Y-W (2019) Transcriptional regulation of carotenoid biosynthesis in plants: so many regulators, so little consensus. Front Plant Sci 10:1017. https://doi.org/10.3389/fpls.2019.01017
Sun T, Yuan H, Cao H, Yazdani M, Tadmor Y, Li L (2018) Carotenoid metabolism in plants: the role of plastids. Mol Plant 11(1):58–74. https://doi.org/10.1016/j.molp.2017.09.010
Takahashi K, Takabayashi A, Tanaka A, Tanaka R (2014) Functional analysis of light-harvesting-like protein 3 (LIL3) and its light-harvesting chlorophyll-binding motif in Arabidopsis. J Biol Chem 289(2):987–999. https://doi.org/10.1074/jbc.M113.525428
Tamura H, Ishikita H (2020) Quenching of singlet oxygen by carotenoids via ultrafast super-exchange dynamics. J Phys Chem A 124(25):5081–5088. https://doi.org/10.1021/acs.jpca.0c02228
Tanaka R, Rothbart M, Oka S, Takabayashi A, Takahashi K, Shibata M, Myouga F, Motohashi R, Shinozaki K, Grimm B (2010) LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc Natl Acad Sci 107(38):16721–16725. https://doi.org/10.1073/pnas.1004699107
Toledo-Ortiz G, Huq E, Rodríguez-Concepción M (2010) Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci 107(25):11626–11631. https://doi.org/10.1073/pnas.0914428107
Tsuyuzaki K, Sato H, Sato K, Nikaido I (2020) Benchmarking principal component analysis for large-scale single-cell RNA-sequencing. Genome Biol 21(1):9. https://doi.org/10.1186/s13059-019-1900-3
Walter MH, Floss DS, Strack D (2010) Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232(1):1–17. https://doi.org/10.1007/s00425-010-1156-3
Wang Y, Wang C, Zheng M, Lyu J, Xu Y, Li X, Niu M, Long W, Wang D, Wang H, Terzaghi W, Wang Y, Wan J (2016) WHITE PANICLE1, a Val-tRNA Synthetase regulating chloroplast ribosome biogenesis in Rice, is essential for early chloroplast development. Plant Physiol 170(4):2110–2123. https://doi.org/10.1104/pp.15.01949
Welsch R, Zhou X, Yuan H, Alvarez D, Sun T, Schlossarek D, Yang Y, Shen G, Zhang H, Rodriguez-Concepcion M, Thannhauser TW, Li L (2018) Clp protease and OR directly control the Proteostasis of Phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol Plant 11(1):149–162. https://doi.org/10.1016/j.molp.2017.11.003
Yokono M, Akimoto S (2018) Energy transfer and distribution in photosystem super/megacomplexes of plants. Curr Opin Biotechnol 54:50–56. https://doi.org/10.1016/j.copbio.2018.01.001
Zapata M, Rodríguez F, Garrido JL (2000) Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar Ecol Prog Ser 195:29–45. https://doi.org/10.3354/meps195029
Zhan W, Liu J, Pan Q, Wang H, Yan S, Li K, Deng M, Li W, Liu N, Kong Q (2019) An allele of Zm PORB 2 encoding a protochlorophyllide oxidoreductase promotes tocopherol accumulation in both leaves and kernels of maize. Plant J 100(1):114–127. https://doi.org/10.1111/tpj.14432
Zhang D, Bao Y, Sun Y, Yang H, Zhao T, Li H, Du C, Jiang J, Li J, Xie L (2020) Comparative transcriptome analysis reveals the response mechanism of Cf-16-mediated resistance to Cladosporium fulvum infection in tomato. BMC Plant Biol 20(1):33. https://doi.org/10.1186/s12870-020-2245-5
Zhou F, Wang C-Y, Gutensohn M, Jiang L, Zhang P, Zhang D, Dudareva N, Lu S (2017) A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proc Natl Acad Sci 114(26):6866–6871
Zhou X, Welsch R, Yang Y, Alvarez D, Riediger M, Yuan H, Fish T, Liu J, Thannhauser TW, Li L (2015a) Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc Natl Acad Sci U S A 112(11):3558–3563. https://doi.org/10.1073/pnas.1420831112
Zhou X, Welsch R, Yang Y, Álvarez D, Riediger M, Yuan H, Fish T, Liu J, Thannhauser TW, Li L (2015b) Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc Natl Acad Sci 112(11):3558–3563. https://doi.org/10.1073/pnas.1420831112
Funding
This work was supported by the Basic and Applied Basic Research Foundation of Guangdong Province (2020A1515110586), the China Postdoctoral Science Foundation (2020 M672839, 2019 M663122, 2020 M672838), the National Key Research and Development Program of China (2017YFE0118100 and 2018YFD02003025), the National Natural Science Foundation of China (31761130073 and 31872169), the Shenzhen Virtual University Park Support Scheme to Shenzhen Research Institute of the Chinese University of Hong Kong and the Hong Kong Research Grant Council (14177617).
Author information
Authors and Affiliations
Contributions
FY, TS and JHZ designed experiments. FY performed experiments. FY, DD and TS analyzed data. FY and DD wrote the manuscript. JHZ and TS critically commented and revised it. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that no competing interests exist.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://dx.doi.org/10.1186/s12284-021-00484-x
Supplementary Information
12284_2021_474_MOESM1_ESM.docx
Additional file 1 Figure S1. OsLIL3 sequence alignment between oslil3 and WT. (a), Sequence alignment between WT and oslil3 16-#1 mutant at DNA level. The red triangle indicates stop codon in the mutated OsLIL3 sequence that leads to premature translational termination while the blue triangle indicates end of truncated sequence due to translational termination. (b), amino acid sequence alignment between mutated oslil3 and WT. Figure S2. (a), Principal component analysis (PCA) was used to describe the overall transcriptome structure in the RNA-seq data. Normalized counts for all genes in the sequenced samples for both mutant and WT samples were projected onto the first two principal components; (b), Hierarchical clustering was used to group 6 main samples (according to the normalized counts of genes in the 3 mutant and 3 WT samples) onto a heatmap shown with its associated dendrogram. Darker colors denote greater similarity between samples. Figure S3. Gene Ontology (GO) enrichment analysis for oslil3 vs WT DEGs. Three types of GO enrichments are included: (a), Biological process; (b), Cellular components; (c), Molecular function. Figure S4. Top 20 enriched KEGG pathways for up and down-regulated DEGs of oslil3 vs WT. Figure S5. Protein-protein interaction between OsLIL3 and OsPSY1 & OsPSY3. (a), Y2H assays. OsLIL3 without chloroplast transit peptide was fused with BD vector. OsPSY1 and OsPSY3 were fused with pGADKT7 vector. The combinations OsLIL3/empty vector (pGADT7), empty vector (pGABDKT7)/OsPSY1 and empty vector (pGBKT7)/OsPSY3 were included as negative controls. Yeast cells containing various vector combinations were grown on DDO plates for co-transformation selection. Yeast cells showing growth and blue color on high selection TDO/X/A plates confirmed positive interactions between the two indicated proteins. (b), OsLIL3-FLAG coimmunoprecipitated with OsPSY1-HA and OsPSY3-HA. Proteins were purified from tobacco leaves expressing OsLIL3-FLAG with OsPSY1-HA and OsPSY3-HA immunoprecipitated with an anti-Flag or anti-HA antibody. The presence of OsLIL3-FLAG, and OsPSY1-HA and OsPSY3-HA in the complex was determined by immunoblot analysis with Flag anti-bodies. (c), BiFC-visualized interactions between OsLIL3 and OsPSY1 and OsPSY3 in N. benthamiana. Abbreviations: YN, YFPN. YC, YFPC. Chl, chlorophyll autofluorescence; YFP, yellow fluorescent protein; BF, bright field. All bars = 5 μm. Figure S6. Y2H assay validation for interaction between OsLIL3 and other proteins involved in carotenoid and chlorophyll biosynthesis whose gene products were identified as DEGs in oslil3 vs WT RNA-seq. Protein sequence without chloroplast transit peptide was fused with Gal DNA-activation domain. The combinations OsLIL3 fused into BD vector and OsPSY2 fused into AD vector were included as positive controls. Yeast cells containing various vector combinations were grown on DDO plates (SD –Leu –Trp) for co-transformation selection. Yeast cells with blue color colony on high selection TDO/X/A plates confirmed positive interactions between the two indicated proteins while no growth signals in TDO/X/A suggest lack of interaction. Figure S7. Alignment and domain comparison of OsLIL3 and AtLIL3.1 & AtLIL3.2 from rice and Arabidopsis. Protein sequences of these proteins were aligned using Jalview 2.0 (Waterhouse et al. 2009). The first transmembrane helix (THM) shows a conserved LHC motif, that is indicated by a black line below the respective sequence while the double black lines show the conserved functional globular domain. Figure S8. Comparison of the fold changes of 15 selected transcripts using two reference genes. Each blue point represents a chosen gene normalized to two reference genes, and the gene name is showed at the end of each linked line.
12284_2021_474_MOESM2_ESM.docx
Additional file 2 Supplementary data S1. Off-target prediction in oslil3 16-#1. Supplementary data S2. Nucleotide mutation in tested oslil3 CRISPR-Cas9 induced mutations. Supplementary data S3. Primers used in this study. Supplementary data S4. Segregation of CRISPR-Cas9 induced mutation in T1 generation. Supplementary data S5. DEGs in ‘Plant-pathogen interaction’ KEGG category. Supplementary data S6. Light harvesting genes in oslil3 vs WT. Supplementary data S7. Cell wall genes list in oslil3 vs WT. Supplementary data S8. Accession number of genes reported in this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, F., Debatosh, D., Song, T. et al. RETRACTED ARTICLE: Light Harvesting-like Protein 3 Interacts with Phytoene Synthase and Is Necessary for Carotenoid and Chlorophyll Biosynthesis in Rice. Rice 14, 32 (2021). https://doi.org/10.1186/s12284-021-00474-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-021-00474-z