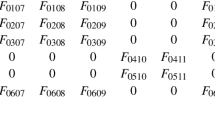Abstract
During the hemostatic phase of wound healing, vascular injury leads to endothelial cell damage, initiation of a coagulation cascade involving platelets, and formation of a fibrin-rich clot. As this cascade culminates, activation of the protease thrombin occurs and soluble fibrinogen is converted into an insoluble polymerized fibrin network. Fibrin polymerization is critical for bleeding cessation and subsequent stages of wound healing. We develop a cooperative enzyme kinetics model for in vitro fibrin matrix polymerization capturing dynamic interactions among fibrinogen, thrombin, fibrin, and intermediate complexes. A tailored parameter subset selection technique is also developed to evaluate parameter identifiability for a representative data curve for fibrin accumulation in a short-duration in vitro polymerization experiment. Our approach is based on systematic analysis of eigenvalues and eigenvectors of the classical information matrix for simulations of accumulating fibrin matrix via optimization based on a least squares objective function. Results demonstrate robustness of our approach in that a significant reduction in objective function cost is achieved relative to a more ad hoc curve-fitting procedure. Capabilities of this approach to integrate non-overlapping subsets of the data to enhance the evaluation of parameter identifiability are also demonstrated. Unidentifiable reaction rate parameters are screened to determine whether individual reactions can be eliminated from the overall system while preserving the low objective cost. These findings demonstrate the high degree of information within a single fibrin accumulation curve, and a tailored model and parameter subset selection approach for improving optimization and reducing model complexity in the context of polymerization experiments.









Similar content being viewed by others
Notes
For example, if \({\mathbf {q}} = [k^{+}_4, k^{-}_2, k^{+}_1, k, k^{+}_2]\) and \(\bar{\mathbf {q}} = [ k^{-}_3 , k^{+}_3 , k^{-}_1 , k^{-} , k^{-}_4 , k^{+}]\), then \(\mathbf {q_k}= \langle {\mathbf {q}},\bar{\mathbf {q}} \rangle = [{\mathbf {q}}[3], \bar{\mathbf {q}}[3], {\mathbf {q}}[5], {\mathbf {q}}[2], \bar{\mathbf {q}}[2], \bar{\mathbf {q}}[1], {\mathbf {q}}[1], \bar{\mathbf {q}}[5], {\mathbf {q}}[4], \bar{\mathbf {q}}[6], \bar{\mathbf {q}}[4]].\) Note that this notation throughout the paper does not denote an inner product.
References
Banks HT, Bekele-Maxwell K, Bociu L, Noorman M, Tillman K (2015) The complex-step method for sensitivity analysis of non-smooth problems arising in biology. Eurasian J Math Comput Appl 3:15–68
Brendel M, Bonvin D, Marquardt W (2006) Incremental identification of kinetic models for homogeneous reaction systems. Chem Eng Sci 61:5404–5420
Briggs GE, Haldane JBS (1925) A note on the kinematics of enzyme action. Biochem J 19:338–339
Brown AC, Baker SR, Douglas AM, Keating M, Alvarez-Elizondo MB, Botvinick EL, Guthold M, Barker TH (2015) Molecular interference of fibrin’s divalent polymerization mechanism enables modulation of multiscale material properties. Biomaterials 49:27–36
Brun R, Kühni M, Siegristm H, Gujer W, Reichert P (2002) Practical identifiability of ASM2d parameters - systematic selection and tuning of parameter sets. Water Res 36:4113–4127
Burth M, Verghese GC, Vélez-Reyes M (1999) Subset selection for improved parameter estimates in on-line identification of a synchronous generator. IEEE Trans Power Syst 14:218–225
Chatterjee MS, Denney WS, Jing H, Diamond SL (2010) Systems biology of coagulation initiation: kinetics of thrombin generation in resting and activated human blood. PLoS Computat Biol 6:e1000950
Chester D, Brown AC (2017) The role of biophysical properties of provisional matrix proteins in wound repair. Matrix Biol 60–61:124–140
Chester D, Marrow EA, Daniele MA, Brown AC (2019) Wound healing and the host response in regenerative engineering. In: Narayan R (ed) Encyclopedia of biomedical engineering. Elsevier, Amsterdam, 1:1-12
Chernysh IN, Nagaswami C, Purohit PK, Weisel JW (2012) Fibrin clots are equilibrium polymers that can be remodeled without proteolytic digestion. Sci Rep 2:879
Cintrón-Arias A, Banks HT, Capaldi A, Lloyd AL (2009) A sensitivity matrix based methodology for inverse problem formulation. J Inverse Ill-posed Prob 17:545–564
Goutelle A, Maurin M, Rougier R, Barbaut X, Bourguignon L, Ducher M, Maire P (2008) The Hill equation: a review of its capabilities in pharmacological modelling. Fundam Clin Pharmacol 22:633–648
Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, Kessenbrock K, Nie Q, Pear WS, Cotsarelis G, Plikus MV (2019) Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat Commun 10(1):650
Haensel D, Jin S, Sun P, Cinco R, Dragan M, Nguyen Q, Cang Z, Gong Y, Vu R, MacLean AL, Kessenbrock K, Gratton E, Nie Q, Dai X (2020) Defining epidermal basal cell states during skin homeostasis and wound healing using single-sell transcriptomics. Cell Rep 30:3932–3947
Holmes MH (2000) Introduct Foundat Appl Math. Springer, New York
Hoffman M, Harger A, Lenkowski A, Hedner U, Roberts HR, Monroe DM (2006) Cutaneous wound healing is impaired in hemophilia B. Blood 108:3053–3060
Janmey PA, Winer JP, Weisel JW (2009) Fibrin gels and their clinical and bioengineering applications. J R Soc Interface 6:1–10
Jorgensen SN, Sanders JR (2016) Mathematical models of wound healing and closure: a comprehensive review. Med Biolog Eng Comput 54:1297–1316
Keener J, Sneyd J (1998) Mathe Physiol. Springer, New York
Lyness J (1967) Numerical algorithms based on the theory of complex variables. In ACM ’67: Proceedings of the 1967 22nd national conference, vol 4, pp 124–134
Lyness J, Moler C (1967) Numerical differentiation of analytic functions. SIAM J Numer Anal 4:202–210
Martins JRRA, Sturdza P, Alonso JJ (2003) The complex-step derivative approximation. ACM Trans Math Softw 29:245–262
Michaelis L, Menten MI (1913) Die kinetik der invertinwirkung. Biochem Z 39:333–369
Nandi S, Brown AC (2017) Characterizing cell migration within three-dimensional in vitro wound environments. J Visual Exp 16:126
Nandi S, Sommerville L, Nellenbach K, Mihalko E, Erb M, Freytes DO, Hoffman M, Monroe D, Brown AC (2020) Platelet-like particles improve fibrin network properties in a hemophilic model of provisional matrix structural defects. J Coll Interface Sci 577:406–418
Nelder J, Mead R (1965) A simplex method for function minimization. Comput J 7:308–313
Quaiser T, Mönnigmann M (2009) System identifiability testing for unambiguous mechanistic modeling - application to JAK-STAT, MAP kinase, and NF-\(\kappa \) B signaling pathway models. BMC Syst Biol 3:50
Rothenberg TJ (1971) Identification in parametric models. Econometrica 39:577–591
Smith R (2013) Uncertain Quantif Theory, Implement Appl. Society for Industrial and Applied Mathematics, Philadelphia
Sproul EP, Hannan R, Brown AC (2018) Characterization of fibrin-based constructs for tissue engineering. In: Chawla K (ed) Biomater Tissue Eng Methods Protocols Methods Molecul Biol. Springer, New York, pp 85–99
Squire W, Trapp G (1998) Using complex variables to estimate derivatives of real functions. SIAM Rev 10:100–112
Tranquillo RT, Murray JD (1992) Continuum model of fibroblast-driven wound contraction: inflammation-mediation. J Theoret Biol 158:135–172
Vajda S, Rabitz H, Walter E, Lecourtier Y (1989) Qualitative and quantitative identifiability analysis of nonlinear chemical kinetic models. Chem Eng Commun 83:191–219
Vallisneri M (2008) Use and abuse of the Fisher information matrix in the assessment of gravitational-wave parameter-estimation prospects. Phys Rev D 77:042001
Valero C, Javierre E, Garcia-Aznar JM, Menzel A, Gomez-Benito MJ (2015) Challenges in the modeling of wound healing mechanisms in soft biological tissues. Ann Biomed Eng 43:1654–1665
Vermolen FJ, Gefen A (2013) A phenomenological model for chemico-mechanically induced cell shape changes during migration and cell-cell contacts. Biomechan Model Mechanobiol 12:301–323
Wang Y, Guerrero-Juarez CF, Qiu Y, Du H, Chen W, Figueroa S, Plikus MV, Nie Q (2019) A multiscale hybrid mathematical model of epidermal-dermal interactions during skin wound healing. ExpDermatol 28:493–502
Weihs D, Gefen A, Vermolen FM (2016) Review on experiment-based two- and three-dimensional models for wound healing. Interface Focus 6:20160038
Weisel JW (2004) The mechanical properties of fibrin for basic scientists and clinicians. Biophys Chem 112:267–276
Weisel JW (2005) Fibrinogen and fibrin. Adv Protein Chem 70:247–299
Weisel JW, Litvinov RI (2013) Mechanisms of fibrin polymerization and clinical implications. Blood 121:1712–1719
Weisel JW, Nagaswami C (1992) Computer modeling of fibrin polymerization kinetics correlated with electron microscope and turbidity observations: clot structure and assembly are kinetically controlled. Biophys J 63:111–128
Wolberg AS (2007) Thrombin generation and fibrin clot structure. Blood Rev 21:124–131
Wolberg AS, Gabriel DA, Hoffman M (2002) Analyzing fibrin clot structure using a microplate reader. Blood Coagulat Fibrinolysis 13:533–539
Funding
This study was funded in part by grants DMS-1638521 and DMR-1847488 (CAREER) from the National Science Foundation and NHLBI R01HL146701 from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pearce, K.J., Nellenbach, K., Smith, R.C. et al. Modeling and Parameter Subset Selection for Fibrin Polymerization Kinetics with Applications to Wound Healing. Bull Math Biol 83, 47 (2021). https://doi.org/10.1007/s11538-021-00876-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-021-00876-6