Abstract
Common genetic variants at the RIN3 locus on chromosome 14q32 predispose to Paget’s disease of bone (PDB) but the mechanisms by which they do so are unknown. Here, we analysed the skeletal phenotype of female mice with targeted inactivation of the mouse Rin3 gene (Rin3−/−) as compared with wild-type littermates. The Rin3−/− mice had higher trabecular bone volume (BV/TV%) compared with wild type. Mean ± standard deviation values at the distal femur at 8 weeks were 9.0 ± 2.5 vs. 7.0 ± 1.5 (p = 0.002) and at 52 weeks were 15.8 ± 9.5 vs. 8.5 ± 4.2 (p = 0.002). No differences were observed in femoral cortical bone parameters with the exception of marrow diameter which was significantly smaller in 52-week-old Rin3−/− mice compared to wild type: (0.43 mm ± 0.1 vs. 0.57 mm ± 0.2 (p = 0.001). Bone histomorphometry showed a lower osteoclast surface / bone surface (Oc.S/BS%) at 8 weeks in Rin3−/− mice compared to wild type (24.1 ± 4.7 vs. 29.7 ± 6.6; p = 0.025) but there were no significant differences in markers of bone formation at this time. At 52 weeks, Oc.S/BS did not differ between genotypes but single labelled perimeter (SL.Pm/B.Pm (%)) was significantly higher in Rin3−/− mice (24.4 ± 6.4 vs. 16.5 ± 3.8, p = 0.003). We conclude that Rin3 negatively regulates trabecular bone mass in mice by inhibiting osteoclastic bone resorption and favouring bone formation. Our observations also suggest that the variants that predispose to PDB in humans probably do so by causing a gain-in-function of RIN3.
Similar content being viewed by others
Introduction
Paget’s disease of bone (PDB) is characterised by increased bone remodelling at one or more skeletal sites. It is thought that the primary pathogenic event in PDB is increased osteoclast activity, but this is accompanied by increased and disorganised bone formation. Genetic factors play a key role in the pathogenesis of PDB, but environmental triggers also play a role as reflected by a reduction in the prevalence and severity of PDB in many countries over recent decades [1]. Genome-wide association studies (GWAS) have so far identified six loci that predispose to PDB at a genome-wide significant level [2, 3], and additional predisposing genes have been identified by linkage analysis and genome sequencing in families [4,5,6]. A locus on chromosome 14q32 was found to predispose to PDB by an extended GWAS [2]. The strongest association was observed with rs10498635 which is situated close to the RIN3 gene, encoding the Ras and Rab interactor protein 3 [7]. The rs10498635 variant lies outside the gene but is in strong linkage disequilibrium with an arginine to cysteine amino acid change at position 279 (R279C) of the protein [2]. Subsequent mutation screening of RIN3 identified several other protein coding variants that were associated with PDB which cluster in a proline-rich domain of the protein [8]. In another study of Belgian PDB patients and controls, de Ridder and colleagues [9] confirmed evidence of association between variants at the RIN3 locus and PDB and identified several rare potentially deleterious coding variants that were associated with the disease. While the role of Ras and Rab interactor 3 in bone metabolism remains unclear, it is a guanine exchange factor for small GTPases which are known play a role in vesicular trafficking and other processes relevant to osteoclastic bone resorption [10, 11]. With this background, the aim of the present study was to gain an insight into the role of RIN3 in skeletal homeostasis by conducting skeletal phenotyping of mice with targeted inactivation of the Rin3 gene.
Materials and Methods
Animals
Mice with targeted inactivation of Rin3 (Rin3−/−), generated on a C57BL/6J background, were obtained from the Mutant Mouse Resource and Research Center (MMRC) catalogue number 049460-UCD, through the Jackson Laboratory. Deletion of Rin3 had been obtained by transfecting embryonic stem cells with the targeting vector containing a Velocigene cassette (ZEN-Ub1). The targeting procedure generated mice with a 29.89Kb deletion at the Rin3 locus encompassing part of exon 1, all of exon 2 and most of exon 3 of Rin3. The Rin3−/− mice were crossed with 129P2OlaHsd mice obtained from Harlan UK to generate Rin3+/− heterozygotes on a mixed C57BL/6J and 129P2OlaHsd genetic background. All experiments were performed on female Rin3−/− and wild-type littermates generated from mating of Rin3+/- heterozygotes. We confirmed that mRNA for Rin3 was undetectable in lung tissue from Rin3−/− mice using qPCR, confirming that the targeted inactivation of Rin3 was successful (data not shown). The mice were housed in a designated animal facility in pathogen-free rooms maintained at constant temperature, with 12-h light/12-h dark cycles. They had free access to water and pelleted standard commercial diet (SDS, Special Diets Service). Mice were sacrificed by cervical dislocation or decapitation according to Schedule 1 of the Animals (Scientific Procedures) Act. The sample size for the in vivo experiment was chosen to provide at least 80% power to detect a 1.3 standard deviation difference in BV/TV between Rin3−/− and wild-type (WT) mice.
Analysis of Rin3 Gene Expression
In order to assess the efficiency of the targeted inactivation procedure, RNA was extracted from homogenised lung tissue of 7.5-week-old mice using TRizol® reagent (Invitrogen). Complementary DNA (cDNA) was transcribed by reverse transcription-polymerase chain reaction (RT-PCR) using the SuperScript III Reverse Transcriptase kit (Invitrogen). The expression Rin3 was assessed by quantitative PCR with the Roche universal library probe system (Roche). A FAM-labelled probe (#13, AGGCAGAG) was used with the following manually designed primers from the Ensembl Genome Browser: forward 5′-GCCGGTCCTATTCCAGATG-3′; Reverse 5′-AAGAACTGAGCCTTCCAGGTA-3′. The reaction was performed using the SensiFAST Probe kit (Bioline) on a Chromo 4TM thermocycler (M J Research). The levels of Rin3 were normalised against eukaryotic 18S ribosomal RNA, which was detected using a VIC-labelled probe-primer (Applied Biosystems).
Transcriptome Analysis by RNA Sequencing
We evaluated global gene expression in bone marrow derived osteoclast cultures and primary calvarial osteoblast cultures derived from Rin3−/− and wild-type mice. The cultures were prepared from three mice of each genotype for each cell type and total RNA was extracted using TRizol® reagent (Invitrogen). The TruSeq Stranded Total RNA library Gold prep kit (Illumina) was used for preparation of the cDNA library and sequencing was performed using an Illumina HiSeq 4000 75PE to yield 290 Million + 290 Million reads per lane in 2 lanes. Full details of the methods used to process the data and to analyse gene expression are provided in the supplementary information.
MicroCT Analysis
Bone density and structure were analysed by micro computed tomography (microCT) using a Skyscan 1172 instrument (Bruker, Belgium). Prior to analysis, the bone samples were dissected free of soft tissue and fixed in 4% (v/v) paraformaldehyde in PBS overnight. The samples were then washed twice in PBS and stored at 4 °C in 70% (v/v) ethanol until scanning. Analysis of trabecular bone was performed by scanning the distal femoral and proximal tibial metaphysis and the 6th lumbar vertebra. At the distal femoral metaphysis, 450 slices were analysed in a region of interest 750 µm from the growth plate. At the proximal tibial metaphysis, 200 slices were analysed in a region of interest 500 µm from the growth plate. At lumbar vertebra, 200 slices were analysed 250 µm distally the transverse process. Analysis of cortical bone was performed on the femoral diaphysis only, where 200 slices were analysed 5 mm from the growth plate. The samples were scanned using an X-Ray source set at 60 kV and 167 µA, a 0.5 mm aluminium filter and a resolution of 5 µm. The cross-section images were reconstructed using the NRecon system (Bruker, Belgium). The transaxial view of this model was then analysed with a free hand tracing tool at the region of interest, using the CTAn system (Bruker, Belgium). Both hind legs were used for the trabecular and cortical analyses.
Bone Histomorphometry
Bone samples were processed and stained for histology using standard methods as previously described [12]. Briefly, animals received intraperitoneal injections of calcein 6 days and 2 days before culling. The skin was removed, the bone samples were fixed for 24 h in 4% paraformaldehyde and stored at 4 °C in 70% ethanol until use. The samples were embedded in methyl methacrylate and 5 µm sections were cut using a diamond knife. For analysis of bone resorption parameters, sections were stained for tartrate resistant acid phosphatase (TRAcP) to visualise osteoclasts and counterstained with aniline blue. For analysis of calcein double labelling, sections were counterstained with calcein blue. The bone sections were analysed using a Zeiss LSM800 (Carl Zeiss Ltd., UK) fluorescence microscope. Histomorphometric measurements were performed using an open source software programme [13] by an observer blinded to genotype.
Biochemical Markers
We measured serum levels of the procollagen I N-terminal propeptide (PINP) and the type I Collagen Cross-Linked C-Telopeptide (CTX-I) by enzyme-linked immunosorbent assay using kits purchased from Immunodiagnostic Systems (IDS) (Boldon UK). Catalogue numbers were AC33F1 (PINP) and AC-06F (CTX-I) and they assays were run according to the manufacturer’s instructions. For both assays, values were recorded based on mean of the duplicate samples and expressed in ng/ml by calibrating against a standard curve.
Osteoclast Cultures
Osteoclasts were generated from bone marrow cells flushed from the long bones of female mice aged between 9 and 14 weeks. The marrow samples were cultured for 2 days in alpha-MEM supplemented with 1% L-Glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 10% foetal calf serum (Hyclone) and 100 ng/ml M-CSF (R&D systems Abingdon UK). Non-adherent cells were removed and the adherent macrophages were plated at 1 × 104 cells in 96-well plates in the presence of 25 ng/ml M-CSF and 75 ng/ml RANKL (R&D Systems) for another 3 days. The medium was changed again in the same conditions and the cells were kept for another 24 h in the incubator. At the end of the culture period, the cells were fixed in 4% paraformaldehyde and stained for TRAcP according to standard techniques. Cells with at least three nuclei that stained positive for TRAcP were counted as osteoclasts. Osteoclasts that contained ≥10 nuclei were considered to be large osteoclasts.
We also studied osteoclast survival in the same cultures. For this assay, RANKL was added at a reduced concentration of 25 ng/ml for the last 4 days of the culture and then the medium was replaced with standard culture medium without RANKL. The cultures continued for up to 72 h from when the cells were fixed, then stained for TRAcP and counted as described above.
Osteoblast Cultures
Primary osteoblasts were isolated from the calvarial bones of 2-day-old mice using sequential collagenase digestion as previously described [14]. The cells were cultured in 75 cm2 flasks in standard alpha-MEM supplemented with 10% foetal bovine serum and left to adhere overnight. To measure the activity of alkaline phosphatase (ALP), the osteoblasts were passaged using trypsin and seeded into 96-well plates at a density of 8 × 103 cells per well. The cells were cultured for 7 days and the experiment was terminated by the addition of lysis buffer containing 1M diethanolamine, 1 mM MgCl2, and 0.05% Triton X100. The ALP activity was assessed on cell lysates using a colorimetric assay based on conversion of p-nitrophenyl phosphate to p-nitrophenol using a plate-reader set at an absorbance of 405 nm. Activity of ALP was corrected for cell number following staining with 10% Alamar blue reagent (v/v) as described [15]. We also assessed formation of mineralised bone nodules using calvarial osteoblast cultures. For this assay, the osteoblasts were plated in 12-well plates at a density of 1 × 105 cells per well and cultured for 21 days in osteogenic media as described [15]. At the end of the culture period, cell viability was assessed using staining with 10% Alamar blue reagent (v/v). The cells were then fixed in 70% ethanol, washed in PBS, briefly stained with alizarin red and left to dry overnight. The nodules were quantified by destaining the cultures in a 10% (w/v) cetylpyridinium chloride solution prepared in 10mM sodium phosphate (pH 7.0) and measuring absorbance of the extracted stain by spectrophotometry as previously described [16]. The absorbance was measured at 562 nm and compared to a standard curve. The values were corrected for cell number as assessed by alamar blue staining. All colorimetric and fluorescent measures were done using the BioTek Synergy HT plate reader (BioTek, UK).
Statistical Analysis
Differences between genotype groups for continuous variables were assessed by t test for independent samples in individual experiments. For the cell culture experiments a general linear model analysis of variance was used to study the effect of time and genotype in which all the replicates within individual experiments were combined. For this analysis, the variable of interest (osteoclast number, alizarin red staining, alkaline phosphatase) was selected as the dependent variable and (where relevant) time, experiment and genotype entered as the explanatory variables. Statistical analysis was performed using SPSS version 22 or Minitab version 18. A p value of less than 0.05 was considered to be statistically significant.
Results
General Appearance and Body Weight
The Rin3−/− mice were of similar appearance to wild-type littermates and had a similar body weight at 8 weeks of age (Rin3−/− vs. WT (mean ± SD) = 20.8 ± 2.4 vs. 21.3 ± 1.5 g, p = 0.59) and 52 weeks of age (34.1 ± 4.6 vs. 33.4 ± 1.5 g, p = 0.60).
MicroCT Analysis
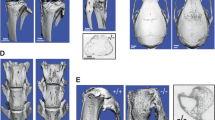
The results of microCT analysis of trabecular bone at the femoral metaphysis in 8-week and 52-week-old mice are shown in Table 1. Trabecular bone volume (BV/TV) and trabecular number were significantly higher at the femoral metaphysis of Rin3−/− mice at 8 weeks of age compared with wild-type littermates. Trabecular thickness did not differ between genotypes, but trabecular separation and trabecular patterning factor were both lower in the Rin3−/− mice as compared with wild type. At 52 weeks, the difference between genotypes was even more pronounced with substantially higher BV/TV in the Rin3−/− mice compared to the wild-type littermates. Similar findings were observed at both 8 weeks and 52 weeks at the proximal tibial metaphysis (supplementary information Table S1). Analysis of cortical bone at the femoral diaphysis showed that there was no significant difference in cortical bone mass or structure between Rin3−/− mice and wild-type littermates, except at 52 weeks where the marrow diameter was significantly smaller in Rin3−/− mice compared with wild-type littermates (Table 2). Representative microCT images from the distal femoral metaphysis and the femoral diaphysis in Rin3−/− mice and wild-type littermates at 8 weeks are shown in Fig. 1. In contrast to findings in the limbs, there were no differences in trabecular bone volume or other parameters at the lumbar vertebra 6 in both age groups of Rin3−/− mice compared with wild type (supplementary information Table S2).
Representative microCT images from Rin3−/− and wild-type mice. Panel a trabecular bone in distal femoral metaphysis from wild-type mice (WT); Panel b trabecular bone from distal femoral metaphysis Rin3−/− mice; Panel c cortical bone from femoral diaphysis in wild-type mice; Panel d: cortical bone from femoral diaphysis in Rin3−/− mice. The microCT images are from 8-week-old mice
Bone Histomorphometry
The results of bone histomorphometry are shown in Table 3. Osteoclast surface expressed as a percentage of bone surface (Os.S/BS) was significantly reduced in 8-week-old Rin3−/− mice compared with wild-type littermates. A similar trend was observed for the number of osteoclasts per unit of bone surface (N.Oc/BS) and for the number of osteoclasts per unit of bone volume (N.Oc/BV), but the differences were not significant for these variables. At 52 weeks, Oc.S/BS, N.Oc/BS and N.Oc/BV were numerically lower in Rin3−/− mice compared wild-type littermates but there was considerable variation and the differences between genotypes was not significant. Mineral apposition rate (MAR) did not significantly differ between genotypes at either 8 weeks or 52 weeks. The perimeter of bone surfaces occupied by single labels (sL.Pm/B.Pm) was significantly higher in Rin3−/− mice at 52 weeks. Representative photomicrographs of trabecular bone from 8-week-old and 52-week-old mice are shown in Fig. 2 which illustrates the difference between genotypes in trabecular bone volume throughout life, the relative reduction in osteoclast surfaces in Rin3−/− mice compared to wild type at 8 weeks of age and the relative increase in single labelled surfaces compared to wild type in Rin3−/− mice at 52 weeks of age.
Representative bone histology from Rin3−/− and wild-type mice at aged 8 weeks panels a–d and 52 weeks (Panels e–h). Calcein labelling (light green) in panels a, b, e and f has been visualised under polarised light. Panels c, d, g and h have been stained with aniline blue and counterstained for tartrate resistant acid phosphatase (red). Higher magnification images of the sections are shown in the bottom left of each panel and the area from which the images are taken are indicated by squares with interrupted lines. The black circles in sections c, d and h are air bubbles in the fixative
Biochemical Markers
We found no significant differences between genotype groups in serum concentrations of either PINP or CTX-I values at either 8 weeks or 52 weeks of age. Circulating concentrations of PINP fell significantly with age between 8 weeks and 52 weeks in both genotype groups (both p < 0.001). Concentrations of CTX-1 also fell with age and this was significant in the Rin3−/− mice (p = 0.03) but not in the wild-type mice (p = 0.45) (Table 4).
Osteoclast and Osteoblast Cultures
We found no differences in M-CSF and RANKL-induced osteoclast formation in cultures prepared from Rin3−/− and wild-type littermates either in terms of total osteoclasts or large osteoclasts with more than ten nuclei (Fig. 3a). There was also no difference between genotype groups in osteoclast survival after withdrawal of RANKL (Fig. 3b). Mineralised nodule formation was slightly but significantly increased in Rin3−/− compared with wild-type mice when data from all experiments were combined (Fig. 4). There was no significant difference in alkaline phosphatase activity in calvarial osteoblasts isolated from Rin3−/− and wild-type mice (data not shown).
Osteoclast formation and survival in Rin3−/− and wild-type mice. Panel a: Osteoclast cultures from wild-type (WT) and Rin3−/− mice, showing total osteoclasts and large osteoclasts (≥ 10 nuclei); Panel b: Osteoclast survival following RANKL withdrawal in wild-type (WT) and Rin3−/− mice. There were no significant differences between genotypes, either in osteoclast formation, formation of large osteoclasts or in osteoclast survival at any timepoint. The columns are least square means and bars are standard deviation of the mean for five separate experiments for osteoclast formation and for three separate experiments for osteoclast survival
Mineralised nodule formation in Rin3−/− and wild-type mice. Panel a Mineralised nodule formation in arbitrary units quantitated by alizarin red staining in cultured calvarial osteoblasts from wild-type (WT) and Rin3−/− mice. The columns are least square means and bars are standard deviations pooled from five separate experiments. *p = 0.02 between genotype groups assessed by GLM ANOVA. Panel b representative photomicrographs from bone nodule cultured after 7, 14 and 21 days
Transcriptome Analysis
Results of the transcriptome analysis assessed by RNA sequencing are summarised in supplementary Fig. S1 and supplementary Table S4. We identified eight transcripts from seven genes that were differentially expressed in calvarial osteoblasts cultured from Rin3−/− and wild-type mice. Only two of the transcripts are known to have a role in bone metabolism and both were upregulated in Rin3−/− mice (Sfrp2 and Dlk1). The SFPR2 gene has been implicated in syndactyly and brachydactyly in humans [17] but the increased levels of expression seem unlikely to contribute to the phenotype observed in Rin3−/− mice. The Dlk1 gene is a member of the Notch family which has been implicated as a paracrine and endocrine regulator of bone mass which inhibits osteoblast differentiation [18]. The upregulation of this gene also seems unlikely to contribute to the skeletal phenotype in Rin3−/− mice. We observed no significant differences in patterns of gene expression in osteoclasts generated from bone marrow macrophages between Rin3−/− and wild-type mice when the data were corrected for multiple testing.
Discussion
The RIN3 gene was identified as a candidate for susceptibility to PDB by an extended genome-wide association study [2]. The strongest association was with rs10498635 [2] which is in strong linkage disequilibrium with rs117068593, a common missense variant that encodes an arginine to cysteine amino acid substitution at codon 297 (p.R279C) in exon 6 of RIN3 [8]. Targeted sequencing of RIN3 subsequently identified thirteen missense variants within the same haplotype block as rs117068593 which, when combined, were strongly over-represented in PDB cases as compared with controls with an odds ratio of greater than 3 [8]. A further study confirmed the association between the common p.R279C variant and PDB in Belgian and Dutch patients and identified six further rare missense variants and PDB [9]. In both studies, in silico analysis indicated that p.R279C and many of the rare missense variants were likely to affect protein function, but it remains unclear at present whether they inhibit or enhance the ability of RIN3 to act as a guanine exchange factor.
The results of the present study are of interest in confirming that RIN3 plays an important role in regulating bone metabolism, further supporting the candidacy of this gene as a cause of PDB. Trabecular bone volume and trabecular numbers were substantially higher in Rin3−/− mice at 8 weeks of age as compared with wild-type littermates and the difference between genotypes was even greater in mice aged 52 weeks. The increase in penetrance of the phenotype with ageing is of interest in relation to the fact that PDB increases in prevalence with age in humans [19]. It is also consistent with observations in the P394L mouse model of PDB where the phenotype worsened with age and was fully penetrant in 52-week-old mice [20]. Histomorphometric analysis revealed that this was associated with significantly lower osteoclast surfaces at 8 weeks and significantly higher perimeter occupied by single calcein labels at 52 weeks. It is important to emphasise, however, that the differences between genotypes in these parameters were modest and there was no difference between genotype groups in osteoclast numbers expressed as a function of bone surface or bone volume, mineral apposition rate, bone formation rate, or double labelled calcein perimeter at either time point. While analysis of biochemical markers of bone resorption and formation at both 8 weeks and 52 weeks did not reveal a significant difference between groups, the microCT data coupled with the histomorphometry data do indicate that deficiency of Rin3 causes a relative uncoupling of bone resorption and formation which operates throughout life to cause a markedly increased trabecular bone mass in older mice.
Although trabecular bone mass was markedly higher in Rin3−/− mice, we observed no difference between genotypes in terms of cortical bone parameters except at 52 weeks where the medullary cavity diameter was smaller in Rin3−/− mice compared with wild type, which would be consistent with lower rates of endosteal bone resorption. The contrasting effects of Rin3 deficiency on trabecular bone phenotype versus cortical bone are unexplained at present, but this finding is not infrequent in mice with targeted inactivation of genes presumably due to site-specific effects of their products on bone metabolism [21,22,23].
We also studied osteoclast differentiation and osteoclast survival in vitro but found no significant differences between genotypes. This is consistent with a model whereby the high bone mass and reduced osteoclast surfaces in Rin3−/− mice were due to a decrease in the amount of bone resorbed per osteoclast rather than by an effect on osteoclast differentiation or survival. The phenotype in Rin3−/− mice is of interest in relation to previous studies by Pavlos and colleagues [24] who conducted phenotyping of mice with targeted inactivation of the small GTPase Rab3D, which is a potential target for RIN3. In this study [24], osteoclast differentiation was normal in vitro, but trabecular bone mass was increased in vivo as reported here and this was accompanied by a reduction in eroded surface. In the Pavlos study, it was also of interest that osteoclasts derived from mice with Rab3D deficiency had a defect in resorptive function which was thought to account for the phenotype observed. We did not have the opportunity to study bone resorption by isolated osteoclasts in Rin3−/− mice which is a limitation of the present study, but this work is being planned in the coming months in order to gain greater insight into the role of Rin3 in osteoclast function and bone metabolism. Given that PDB is characterised by increased osteoclast activity and bone resorption, the phenotype we observed in Rin3−/− mice suggests that the missense genetic variants in RIN3 that are associated with PDB may exert gain-of-function effects on bone resorption although more work will be required to confirm this.
Until now, the role of Rin3 in bone metabolism has been little studied. It is recognised that Rin3 acts as a guanine exchange factor (GEF) for several members of the Rab5 subfamily of GTPases [7, 25, 26]. The Rab proteins play an important role in membrane trafficking in several cell types, and studies by Zhao and colleagues have shown that osteoclasts express many different Rab proteins [10]. There is experimental evidence to show that Rab3D [24] and Rab7 [27] both play a role in osteoclastic bone resorption, but it remains unclear if Rin3 acts as a GEF for these Rab family members. It is similarly unclear to what extent the Rab5 subfamily members regulate osteoclast function and further work will be required to investigate the targets for Rin3 in osteoclasts and the mechanisms by which it affects osteoclast function.
The studies reported here are also of interest in relation to the work of Kemp and co-workers who identified RIN3 as a significant determinant of lower-limb BMD in children [28]. In Kemp’s study, higher BMD values in the lower limbs were associated with the rs754388 variant at genome-wide significant level. The rs754388 variant is in strong linkage disequilibrium with the same direction of effect as the variants that we previously reported to be associated with PDB [8]. At first sight, it seems counter-intuitive that variants associated with a condition characterised by increased osteoclast activity might also be associated with high bone mass in children, but it is possible to speculate that positive coupling between bone resorption and formation during the high turnover state that is characteristic of a growing skeleton could account for this finding. In this regard, variants at the RIN3 locus do not appear to be associated with BMD as assessed by DEXA in adults [29] or with heel ultrasound properties of bone [30] at a genome-wide significant level.
In conclusion, the present studies confirm that Rin3 regulates bone mass and bone metabolism in mice with effects on cells of both the osteoclast and the osteoblast lineage, confirming the importance of guanine exchange factors and their targets in regulation of skeletal homeostasis. Our observations also suggest that the variants that predispose to Paget’s disease of bone in humans probably do so by causing a gain-in-function of RIN3, but further work will need to be performed to define the molecular mechanisms by which Rin3 affects bone cell activity.
Change history
19 April 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00223-021-00849-w
References
Corral-Gudino L, Borao-Cengotita-Bengoa M, Del Pino-Montes J, Ralston SH (2013) Epidemiology of paget’s disease of bone: a systematic review and meta-analysis of secular changes. Bone 55:347–352
Albagha OME, Wani S, Visconti MR, Alonso N, Goodman K, Cundy T, Brandi ML, Chung PY, Dargie R, Devogelaer JP, Falchetti A, Fraser WD, Gennari L, Gianfrancesco F, Hooper MJ, Van Hul W, Isaia G, Nicholson GC, Nuti R, Del Pino MJ, Ratajczak T, Rea SL, Rendina D, Gonzalez-Sarmiento R, Di SM, Ward L, Walsh JP, Ralston SH (2011) Genome-wide association identifies three new susceptibility loci for Paget’s disease of bone. Nat Genet 43:685–689
Albagha OM, Visconti MR, Alonso N, Langston AL, Cundy T, Dargie R, Dunlop MG, Fraser WD, Hooper MJ, Isaia G, Nicholson GC, Del Pino MJ, Gonzalez-Sarmiento R, Di SM, Tenesa A, Walsh JP, Ralston SH (2010) Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nat Genet 42:520–524
Ralston SH, Taylor JP (2019) Rare inherited forms of paget’s disease and related syndromes. Calcif Tissue Int 104:501–516
Scotto di Carlo F, Pazzaglia L, Esposito T, Gianfrancesco F (2020) The loss of profilin 1 causes early onset paget’s disease of bone. J Bone Miner Res 35:1387–1398
Divisato G, Formicola D, Esposito T, Merlotti D, Pazzaglia L, Del FA, Siris E, Orcel P, Brown JP, Nuti R, Strazzullo P, Benassi MS, Cancela ML, Michou L, Rendina D, Gennari L, Gianfrancesco F (2016) ZNF687 mutations in severe paget disease of bone associated with giant cell tumor. Am J Hum Genet 98:275–286
Kajiho H, Saito K, Tsujita K, Kontani K, Araki Y, Kurosu H, Katada T (2003) RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J Cell Sci 116:4159–4168
Vallet M, Soares DC, Wani S, Sophocleous A, Warner J, Salter DM, Ralston SH, Albagha OM (2015) Targeted sequencing of the Paget’s disease associated 14q32 locus identifies several missense coding variants in RIN3 that predispose to Paget’s disease of bone. Hum Mol Genet 24:3286–3295
De Ridder R, Boudin E, Vandeweyer G, Devogelaer JP, Fransen E, Mortier G, Van Hul W (2019) Genetic variation in RIN3 in the belgian population supports its involvement in the pathogenesis of paget’s disease of bone and modifies the age of onset. Calcif Tissue Int 104:613–621
Zhao H, Ettala O, Vaananen HK (2002) Intracellular membrane trafficking pathways in bone-resorbing osteoclasts revealed by cloning and subcellular localization studies of small GTP-binding rab proteins. Biochem Biophys Res Commun 293:1060–1065
Itzstein C, Coxon FP, Rogers MJ (2011) The regulation of osteoclast function and bone resorption by small GTPases. Small GTPases 2:117–130
Erben RG, Glosmann M (2019) Histomorphometry in Rodents. Methods Mol Biol 1914:411–435
van ’t Hof RJ, Rose L, Bassonga E, Daroszewska A (2017) Open source software for semi-automated histomorphometry of bone resorption and formation parameters. Bone 99:69–79
Sophocleous A, Landao-Bassonga E, van ’t Hof RJ, Idris AI, Ralston SH (2011) The type 2 cannabinoid receptor regulates bone mass and ovariectomy-induced bone loss by affecting osteoblast differentiation and bone formation. Endocrinology 152:2141–2149
Idris AI, Sophocleous A, Landao-Bassonga E, Canals M, Milligan G, Baker D, van ’t Hof RJ, Ralston SH (2009) Cannabinoid receptor type 1 protects against age-related osteoporosis by regulating osteoblast and adipocyte differentiation in marrow stromal cells. Cell Metab 10:139–147
Stewart TL, Roschger P, Misof BM, Mann V, Fratzl P, Klaushofer K, Aspden RM, Ralston SH (2005) Association of COLIA1 Sp1 alleles with defective bone nodule formation in vitro and abnormal bone mineralisation in vivo. Calcified Tissue Int 77:113–118
Morello R, Bertin TK, Schlaubitz S, Shaw CA, Kakuru S, Munivez E, Hermanns P, Chen Y, Zabel B, Lee B (2008) Brachy-syndactyly caused by loss of Sfrp2 function. J Cell Physiol 217:127–137
Abdallah BM, Ditzel N, Mahmood A, Isa A, Traustadottir GA, Schilling AF, Ruiz-Hidalgo MJ, Laborda J, Amling M, Kassem M (2011) DLK1 is a novel regulator of bone mass that mediates estrogen deficiency-induced bone loss in mice. J Bone Miner Res 26:1457–1471
van Staa TP, Selby P, Leufkens HG, Lyles K, Sprafka JM, Cooper C (2002) Incidence and natural history of Paget’s disease of bone in England and Wales. J. Bone Miner. Res 17:465–471
Daroszewska A, van ’t Hof RJ, Rojas JA, Layfield R, Landao-Basonga E, Rose L, Rose K, Ralston SH (2011) A point mutation in the ubiquitin-associated domain of SQSMT1 is sufficient to cause a Paget’s disease-like disorder in mice. Hum Mol Genet 20:2734–2744
Sophocleous A, Idris AI, Ralston SH (2014) Genetic background modifies the effects of type 2 cannabinoid receptor deficiency on bone mass and bone turnover. Calcif Tissue Int 94:259–268
Windahl SH, Borjesson AE, Farman HH, Engdahl C, Moverare-Skrtic S, Sjogren K, Lagerquist MK, Kindblom JM, Koskela A, Tuukkanen J, Divieti PP, Feng JQ, Hlman-Wright K, Antonson P, Gustafsson JA, Ohlsson C (2013) Estrogen receptor-alpha in osteocytes is important for trabecular bone formation in male mice. Proc Natl Acad Sci USA 110:2294–2299
Moverare-Skrtic S, Wu J, Henning P, Gustafsson KL, Sjogren K, Windahl SH, Koskela A, Tuukkanen J, Borjesson AE, Lagerquist MK, Lerner UH, Zhang FP, Gustafsson JA, Poutanen M, Ohlsson C (2015) The bone-sparing effects of estrogen and WNT16 are independent of each other. Proc Natl Acad Sci USA 112:14972–14977
Pavlos NJ, Xu J, Riedel D, Yeoh JS, Teitelbaum SL, Papadimitriou JM, Jahn R, Ross FP, Zheng MH (2005) Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption. Mol Cell Biol 25:5253–5269
Kajiho H, Sakurai K, Minoda T, Yoshikawa M, Nakagawa S, Fukushima S, Kontani K, Katada T (2011) Characterization of RIN3 as a guanine nucleotide exchange factor for the Rab5 subfamily GTPase Rab31. J Biol Chem 286:24364–24373
Yoshikawa M, Kajiho H, Sakurai K, Minoda T, Nakagawa S, Kontani K, Katada T (2008) Tyr-phosphorylation signals translocate RIN3, the small GTPase Rab5-GEF, to early endocytic vesicles. Biochem. Biophys. Res. Commun 372:168–172
Zhao H, Laitala-Leinonen T, Parikka V, Vaananen HK (2001) Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J Biol Chem 276:39295–39302
Kemp JP, Medina-Gomez C, Estrada K, St Pourcain B, Heppe DH, Warrington NM, Oei L, Ring SM, Kruithof CJ, Timpson NJ, Wolber LE, Reppe S, Gautvik K, Grundberg E, Ge B, van der Eerden B, van de Peppel J, Hibbs MA, Ackert-Bicknell CL, Choi K, Koller DL, Econs MJ, Williams FM, Foroud T, Zillikens MC, Ohlsson C, Hofman A, Uitterlinden AG, Davey Smith G, Jaddoe VW, Tobias JH, Rivadeneira F, Evans DM (2014) Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet 10:e1004423
Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, LaCroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, Gonzalez-Macias J, Kahonen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren O, Lorenc RS, Marc J, Mellstrom D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NL et al (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44:491–501
Kemp JP, Morris JA, Medina-Gomez C, Forgetta V, Warrington NM, Youlten SE, Zheng J, Gregson CL, Grundberg E, Trajanoska K, Logan JG, Pollard AS, Sparkes PC, Ghirardello EJ, Allen R, Leitch VD, Butterfield NC, Komla-Ebri D, Adoum AT, Curry KF, White JK, Kussy F, Greenlaw KM, Xu C, Harvey NC, Cooper C, Adams DJ, Greenwood CMT, Maurano MT, Kaptoge S, Rivadeneira F, Tobias JH, Croucher PI, Ackert-Bicknell CL, Bassett JHD, Williams GR, Richards JB, Evans DM (2017) Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet 49:1468–1475
Funding
These studies were supported in part by grants to OMEA (311723-GENEPAD) and SHR (787720 – Paget Advance) from the European Research Council and a grant to SHR from Arthritis Research UK (19799).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Mahéva Vallet, Antonia Sophocleous, Anna E. Törnqvist, Asim Azfer, Rob van’t Hof, Omar ME Albagha, Stuart H. Ralston declare that they have no conflicts of interest.
Human and Animal Rights
All animal experiments were approved by the Animal Welfare and Ethical Review Body of the University of Edinburgh and were conducted in accordance with the UK Animals (Scientific Procedures) Act 1986. (Project Licence P2B36BCB0).
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vallet, M., Sophocleous, A., Törnqvist, A.E. et al. Targeted Inactivation of Rin3 Increases Trabecular Bone Mass by Reducing Bone Resorption and Favouring Bone Formation. Calcif Tissue Int 109, 92–102 (2021). https://doi.org/10.1007/s00223-021-00827-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00223-021-00827-2