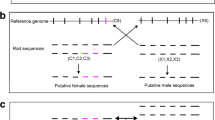
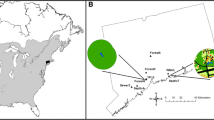
Abstract
Conservation genomics research often relies on accurate sex information to make inferences about species demography, dispersal, and population structure. However, field determined sex data are not always available and can be subject to human error, while laboratory sex assignment methods such as PCR assays can often be costly and challenging for non-model species. Conservation genomics programs increasingly use reduced-representation genome sequencing to assess neutral and functional genetic diversity, population structure, gene flow and pedigrees in threatened species. Here we demonstrate that sex can be determined from reduced-representation sequencing data produced by the increasingly popular Diversity Arrays Technology sequencing workflow (DArT-seq) using a program originally designed for application to shotgun data. This program—sexassign—compares the “dosage” of sequencing reads mapping to autosomes versus the X chromosome. In the present study, sexassign was used to identify the sex of 60 field-collected Greater Stick-Nest Rat (Leporillus conditor) samples, despite the absence of an annotated reference genome for the species. This “read-dosage” approach is not only more accurate and affordable than traditional sex assignment methods, but can be applied to any diploid organism with a heterogametic sex determination system—including non-model and understudied species of conservation importance—by using FASTQs generated by DArT.


Similar content being viewed by others
Data availability
The reads generated for this study have been deposited at the Sequence Read Archive (NCBI) with BioProject ID PRJNA702840 (http://www.ncbi.nlm.nih.gov/bioproject/702840).
Code availability
The original code can be found on Dr Graham Gower’s GitHub repository https://github.com/grahamgower/sexassign.
References
Akane A, Seki S, Shiono H, Nakamura H, Hasegawa M, Kagawa M, Matsubara K, Nakahori Y, Nagafuchi S, Nakagome Y (1992) Sex determination of forensic samples by dual PCR amplification of an X-Y homologous gene. Forensic Sci Int 52(2):143–148. https://doi.org/10.1016/0379-0738(92)90102-3
Barclay SD, Costello B, Sherwin WB (2006) Limited cross-species microsatellite amplification and the isolation and characterization of new microsatellite markers for the greater stick-nest rat (Leporillus conditor). Mol Ecol Notes 6(3):882–885
Bover P, Llamas B, Thomson VA, Pons J, Cooper A, Mitchell KJ (2018) Molecular resolution to a morphological controversy: the case of North American fossil muskoxen bootherium and symbos. Mol Phylogenet Evol 129:70–76. https://doi.org/10.1016/j.ympev.2018.08.008
Clapcote SJ, Roder JC (2005) Simplex PCR assay for sex determination in mice. Biotechniques 38(5):702–706. https://doi.org/10.2144/05385BM05
Cummins D, Kennington WJ, Rudin-Bitterli T, Mitchell NJ (2019) A genome-wide search for local adaptation in a terrestrial-breeding frog reveals vulnerability to climate change. Glob Change Biol 25(9):3151–3162. https://doi.org/10.1111/gcb.14703
Ewart KM, Johnson RN, Ogden R, Joseph L, Frankham GJ, Lo N (2019) Museum specimens provide reliable SNP data for population genomic analysis of a widely distributed but threatened cockatoo species. Mol Ecol Resour 19(6):1578–1592. https://doi.org/10.1111/1755-0998.13082
Flamingh de A, Coutu A, Roca AL, Malhi RS (2020) Accurate sex identification of ancient elephant and other animal remains using low-coverage DNA shotgun sequencing data. Genes Genom Genet 10(4):1427–1432. https://doi.org/10.1534/g3.119.400833
Fowler BLS, Buonaccorsi VP (2016) Genomic characterization of sex-identification markers in Sebastes carnatus and Sebastes chrysomelas rockfishes. Mol Ecol 25(10):2165–2175. https://doi.org/10.1111/mec.13594
Gamble T (2016) Using RAD-seq to recognize sex-specific markers and sex chromosome systems. Mol Ecol 25(10):2114–2116. https://doi.org/10.1111/mec.13648
Gowe G (2019) Inferring the charactersitics of ancient populations using bioinformatic analysis of genome-wide DNA sequencing data [doctoral dissertation]. University of Adelaide
Gower G, Fenderson LE, Salis AT, Helgen KM, van Loenen AL, Heiniger H, Hofman-Kamińska E, Kowalczyk R, Mitchell KJ, Llamas B, Cooper A (2019) Widespread male sex bias in mammal fossil and museum collections. Proc Natl Acad Sci 116(38):19019–19024. https://doi.org/10.1073/pnas.1903275116
Hoffmann A, Decher J, Rovero F, Schaer J, Voigt C, Wibbelt G (2010) Field methods and techniques for monitoring mammals. Man Field Rec Tech Protoc All Taxa Biodivers Invent 8:482–529
Hrovatin K, Kunej T (2017) Genetic sex determination assays in 53 mammalian species: literature analysis and guidelines for reporting standardization. Ecol Evol 8(2):1009–1018. https://doi.org/10.1002/ece3.3707
Jacques M-E, McBee K, Elmore D (2015) Determining sex and reproductive status of rodents. Cooperative Extension Service, Oklahoma
Janečka JE, Davis BW, Ghosh S, Paria N, Das PJ, Orlando L, Schubert M, Nielsen MK, Stout TAE, Brashear W, Li G, Johnson CD, Metz RP, Zadjali AMA, Love CC, Varner DD, Bellott DW, Murphy WJ, Chowdhary BP, Raudsepp T (2018) Horse Y chromosome assembly displays unique evolutionary features and putative stallion fertility genes. Nat Commun 9(1):2945. https://doi.org/10.1038/s41467-018-05290-6
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C, Aschenbrenner-Kilian M, Evers M, Peng K, Cayla C, Hok P, Uszynski G (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. In: Pompanon F, Bonin A (eds) Data production and analysis in population genomics: methods and protocols. Humana Press, Totowa, pp 67–89. https://doi.org/10.1007/978-1-61779-870-2_5
Labov JB, William Huck U, Vaswani P, Lisk RD (1986) Sex ratio manipulation and decreased growth of male offspring of undernourished golden hamsters (Mesocricetus auratus). Behav Ecol Sociobiol 18(4):241–249. https://doi.org/10.1007/BF00300000
Lambert MR, Skelly DK, Ezaz T (2016) Sex-linked markers in the North American green frog (Rana clamitans) developed using DArTseq provide early insight into sex chromosome evolution. BMC Genom 17(1):844. https://doi.org/10.1186/s12864-016-3209-x
McFarlane L, Truong V, Palmer JS, Wilhelm D (2013) Novel PCR assay for determining the genetic sex of mice. Sex Dev 7(4):207–211. https://doi.org/10.1159/000348677
Motahari AS, Bresler G, Tse DNC (2013) Information Theory of DNA shotgun sequencing. IEEE Trans Inf Theory 59(10):6273–6289. https://doi.org/10.1109/TIT.2013.2270273
Pazmiño DA, Maes GE, Green ME, Simpfendorfer CA, Hoyos-Padilla EM, Duffy CJA, Meyer CG, Kerwath SE, Salinas-de-León P, van Herwerden L (2018) Strong trans-Pacific break and local conservation units in the Galapagos shark (Carcharhinus galapagensis) revealed by genome-wide cytonuclear markers. Heredity 120(5):407–421. https://doi.org/10.1038/s41437-017-0025-2
Pečnerová P, Díez-del-Molino D, Dussex N, Feuerborn T, von Seth J, van der Plicht J, Nikolskiy P, Tikhonov A, Vartanyan S, Dalén L (2017) Genome-based sexing provides clues about behavior and social structure in the woolly mammoth. Curr Biol 27(22):3505-3510.e3. https://doi.org/10.1016/j.cub.2017.09.064
Rosenfeld CS, Grimm KM, Livingston KA, Brokman AM, Lamberson WE, Roberts RM (2003) Striking variation in the sex ratio of pups born to mice according to whether maternal diet is high in fat or carbohydrate. Proc Natl Acad Sci 100(8):4628–4632. https://doi.org/10.1073/pnas.0330808100
Sansaloni C, Petroli C, Jaccoud D, Carling J, Detering F, Grattapaglia D, Kilian A (2011) Diversity arrays technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc 5(7):1–2. https://doi.org/10.1186/1753-6561-5-S7-P54
Schultz AJ, Cristescu RH, Littleford-Colquhoun BL, Jaccoud D, Frère CH (2018) Fresh is best: accurate SNP genotyping from koala scats. Ecol Evol 8(6):3139–3151. https://doi.org/10.1002/ece3.3765
Skoglund P, Storå J, Götherström A, Jakobsson M (2013) Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Archaeol Sci 40(12):4477–4482. https://doi.org/10.1016/j.jas.2013.07.004
Steppan SJ, Schenk JJ (2017) Muroid rodent phylogenetics: 900-species tree reveals increasing diversification rates. PLoS ONE 12(8):e0183070. https://doi.org/10.1371/journal.pone.0183070
van Deventer R, Rhode C, Marx M, Roodt-Wilding R (2020) The development of genome-wide single nucleotide polymorphisms in blue wildebeest using the DArTseq platform. Genomics 112(5):3455–3464. https://doi.org/10.1016/j.ygeno.2020.04.032
Acknowledgements
The authors wish to acknowledge and thank Dr Katherine Moseby, Shaun Barclay, Professor Bill Sherwin, and the staff of Arid Recovery Reserve for supplying the field data and samples used in this study.
Funding
This research was supported by the University of Adelaide and funded by the following organisations and awards; Australian Government Research Training Program Scholarship, Nature Foundation South Australia Grand Start Grant (Grant No. 2019–07), Biological Society South Australia/Nature Conservation Society of South Australia Conservation Biology Grant, Field Naturalists Society of South Australia Lirabenda Endowment Fund Research Grant.
Author information
Authors and Affiliations
Contributions
IRO and JJA coordinated submission of samples to DArT. IRO and KJM analysed the data. IRO drafted the abstract, introduction, results, and discussion. KJM drafted the materials and methods and figures. All authors contributed to the interpretation of results and provided feedback on the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Live animal trapping and sampling at Arid Recovery was conducted under South Australian Wildlife Ethics Committee permit numbers 27/98, 4/99, 22/99, 2/2000, 19/2000, and 18/2000.
Consent to publish
The authors give consent for the publication of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Onley, I.R., Austin, J.J. & Mitchell, K.J. Sex assignment in a non-model organism in the absence of field records using Diversity Arrays Technology (DArT) data. Conservation Genet Resour 13, 255–260 (2021). https://doi.org/10.1007/s12686-021-01203-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-021-01203-w