Abstract
• Key message
Parametric indirect models derived from stem analysis of dominant trees were more robust than rule-based machine learning techniques for predicting Site Index of Scots pine stands as a function of climate.
• Context
The uncertainties derived from climate change make it necessary to develop new methods for representing the relationships between site conditions and forest growth.
• Aims
To compare parametric vs nonparametric approaches for modeling site index (SI) of Scots pine stands using bioclimatic variables.
• Methods
We used Random Forest, Boosted Trees, and Cubist techniques for directly predicting the SI of 41 research plots of Scots pine stands, and six parametric models for indirectly predicting SI using stem analysis data. As predictors, we used raster maps of 19 bioclimatic variables.
• Results
The fitted models explained up to \(\sim\)80% of the SI variability, using from five to nine bioclimatic predictors. Though the apparent performance of the parametric models was lower than the rule-based, their bootstrap validation statistics were noticeably higher.
• Conclusion
Parametric indirect models seemed to be the most robust modeling alternative.
Similar content being viewed by others
1 Introduction
Climate change is presumed to cause a significant impact on forest ecosystems during the XXI century (Kirilenko and Sedjo 2007; Lindner et al. 2008). In recent years, the uncertainties derived from forest growth prediction under climate change have given rise to an intense scientific production. In this regard, the development of growth–environment relationships is a preferred standpoint for adapting traditional empirical growth indicators to a changing climate (Fontes et al. 2010). As site index (SI) is the most popular growth indicator for even-aged forest management (Skovsgaard and Vanclay 2008), the development of site index-environment models has become a general research goal (Wang et al. 2004; Seynave et al. 2005; Monserud et al. 2006).
The models developed for this purpose are usually based on the “direct” prediction of SI as a function of climatic, edaphic and/or physiographic predictors by means of a certain regression technique. Admittedly, these models have been increasingly relying on machine learning approaches, which are becoming an attractive modeling alternative since: (1) they provide methods for automatic variable selection, (2) they usually allow for automatically capture nonlinear response–predictor relationships, and (3) some of them allow for automatically model interactions between predictors (González-Rodríguez and Diéguez-Aranda 2020). Reasons (2) and (3) become especially notable when we consider nonparametric learning techniques, such as the “rule-based” approaches, which have become a frequent resource for SI-environment modeling (Crookstion et al. 2010; Barrio-Anta et al. 2020; Watt et al. 2021).
However, as noted by (Sabatia and Burkhart 2014), there is significant controversy regarding the robustness and interpretability of the rule-based models. Concerning robustness, the high amount of parameters and the model complexity implied by the “nonparametric” approach can be relevant sources of overfitting. Additionally, several authors (Weiskittel et al. 2011; Sabatia and Burkhart 2014) found a significant trend to the observational mean in the predicted values resulting from rule-based SI-environment models. These results could be denoting a low extrapolability power for these approaches, which may be a concerning drawback for studies that aim at producing cartographic outputs of forest productivity at regional scales. Concerning interpretability, the model complexity of rule-based approaches may make response–predictor relationships hard to interpret (Aertsen et al. 2010). In this context, using these models for prediction without an adequate interpretation may lead to ecological inconsistencies regarding the theoretical basis of tree growth. Because of these potential disadvantages of rule-based models, parametric approaches that provide similar modeling benefits (automatic variable selection, nonlinear response–predictor relationships and interactions between predictors) have become an attractive line of research in growth–environment relationships modeling (Watt et al. 2015, 2016; Zhu et al. 2019).
Moreover, the observed underperformance of many SI-environment models, including rule-based approaches, might not be entirely due to shortcomings in the chosen regression techniques but also to potential inconsistencies between SI and the environmental predictors used (as suggested by Bontemps and Bouriaud 2014). These potential inconsistencies might be revealing that the “direct” modeling of SI is not really the optimal approach for linking biophysical site attributes with the underlying ecophysiological processes of tree growth. The reason for this might be that the traditional primacy of SI as productivity indicator in even-aged forestry responds exclusively to practical reasons (Skovsgaard and Vanclay 2008), as it lacks any direct ecological meaning. For overcoming the uncertainties of predicting SI, some studies have successfully modeled it in an “indirect” way, usually relying on other growth indicators or parameters that have a more sounding ecological background. For instance, Swenson et al. (2005) mapped the SI of Douglas-fir forests in the USA as a function of the 3-PG process-based model outputs. In this regard, it is important to note that traditional height growth equations (Hossfeld 1822; Gompertz 1825; Richards 1959) already provide nonlinear parametric forms for describing growth processes over time, which are ecologically meaningful as they rely on theoretical assumptions regarding population dynamics and metabolic ecology. As these nonlinear functions are mainly driven by a certain set of growth parameters, it is reasonable to think that these parameters are significantly correlated with the environmental factors that determine tree growth. Considering that SI is an immediate corollary of any age-dependent height growth equation, analyzing the relationships between climate and these growth parameters might be a consistent approach for “indirect” SI-environment modeling.
Our primary objective in this study was to test whether SI could be effectively predicted using parametric approaches that relate ecologically meaningful parameters from growth equations with climatic factors. For testing this, we compared two different ways of predicting SI of Scots pine stands in the northwest of Spain. On the one hand, we performed a direct SI prediction using rule-based models, in a similar way to previous studies (Weiskittel et al. 2011; Sabatia and Burkhart 2014; Barrio-Anta et al. 2020). On the other hand, we proposed a new method for developing SI-environment models by combining simple parametric models with the nonlinear assumptions behind the Hossfeld growth equation.
2 Materials and methods
2.1 Stand height data
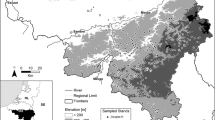
The source of height growth data was a network of permanent research plots established by the Sustainable Forest Management Unit (UXFS) of the University of Santiago de Compostela, Spain. Overall, these plots correspond to pure, even-aged stands of Scots pine (Pinus sylvestris L.) located in the provinces of Lugo and Ourense, in the region of Galicia, and consisted of plantations in communal forests mainly located in mountainous sites (Fig. 1). For this study, we used only the first measurements of the network, carried out in 1996-1997, in which stem analyses of dominant trees was carried out for a set 41 of the plots (González-Rodríguez and Diéguez-Aranda 2021).
Diameter at breast height (dbh) of all trees and total height were measured in each plot. Core samples were also bored in order to count the growth rings. The stand age (t), the number of stems in a hectare (N, trees/ha), the basal area (G, m\(^2\)/ha) and the dominant height (H, m; mean height of the 100 largest-dbh trees per hectare) were calculated from the previous measurements. A summary of the stand variables of the 41 plots considered in this study is shown in Table 1.
For each plot, the SI was estimated using the algebraic difference equation developed by Dieguez-Aranda et al. (2006) for this region, which is based on the Hossfeld growth function (Hossfeld 1822). This was done by projecting the observed dominant height (H) at the measurement age (t) to the reference age of the species (\(t_{ref}\) = 40 years):
being \(H_{2}\) the site index when \(t_2\) = \(t_{ref}\), \(H_1\) = H and \(t_1\) = t.
Stem analyses were carried out following the same procedure described by Dieguez-Aranda et al. (2005a, b). In each of these plots, a subset of two dominant trees per plot (i.e., a total of 82 trees), with heights and diameters differing, respectively, less than 5% to H and D (dominant diameter of the stand), were destructively sampled. Then, five to ten stem slices per tree were extracted along the trees’ height in order to count the growth rings. This allowed for relating tree heights (h) with ages (t), which were corrected using the method proposed by Carmean (1972) and modified by Newberry (1991). From the h-t dataset obtained through stem analysis, we fitted the Hossfeld growth equation for each plot:
being a, b and c the growth parameters for each plot.
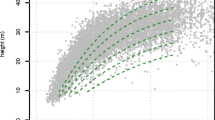
Once these basic parameters were estimated, we calculated another six growth-related derived parameters from Eq. 2: the age, height and growth (\(t_{ip}\), \(h_{ip}\), \(g_{ip}\)) at the inflexion point of the h-t curve; and the age, height and growth (\(t_{mmg}\), \(h_{mmg}\), mmg) at the point of maximum mean growth in height. We selected both points in the growth curve because of their potential eco-physiological significance. An example of a fitted growth curve with inflexion and maximum mean growth points represented is shown in Fig. 2. The inflexion point is reached at the age of maximum growth rate, and corresponds to the second derivative of (2). It represents the turning point between “juvenile” growth and “mature” growth. The maximum mean growth occurs at the age when \(\frac{d}{dt}\left( \frac{h(t)}{t}\right) =0\), and represents the moment from which the height/age ratio starts decreasing. The values and the specific methods to calculate each one of the nine alternative growth parameters estimated are summarized in Table 2.
2.2 Climatic data
As a source of climatic data, we used the Worldclim 2 bioclimatic dataset (Fick and Hijmans 2017). This dataset consists of a collection of raster maps with a spatial resolution of 1 km of climatic historical means for the period 1970-2000. The variables included in the maps are 19 bioclimatic indicators often used in species distribution modeling because of its biological significance. Some of them represent annual trends, such as the annual precipitation (BIO12), whereas others represent differences between seasons, such as the temperature seasonality (BIO3), or extreme climatic events, such as the minimum temperature of the coldest month (BIO6). From these raster maps, we extracted the values of the 19 bioclimatic indicators corresponding to the 41 inventory plot locations, so each site was characterized by a SI value and a set of potential climatic predictors of forest productivity. A summary of the 19 bioclimatic variables proposed as potential predictors of SI is shown in Table 3.
2.3 SI direct prediction
We used three different rule-based learning techniques for directly relating SI to the bioclimatic predictors: Random Forest, Boosted Trees and Cubist. The first and second approaches have been successfully used for SI prediction in other studies (Aertsen et al. 2010; Weiskittel et al. 2011; Sabatia and Burkhart 2014). In contrast, to our knowledge, the Cubist algorithm has never been used before in forest growth modeling. In order to provide a methodological background, we present hereunder a description of the basics and used procedures for fitting these techniques in the current study.
2.3.1 Random forest regression
Random Forest (Breiman 2001) is a rule-based ensemble technique that performs a bagging procedure for developing an unbiased collection of regression trees. At each split of each regression tree, a randomly selected subset of the predictors is used for defining the node, thus granting a unique structure to each tree and avoiding between-tree correlation. This technique has been previously used for SI-environment modeling in other studies (Weiskittel et al. 2011; Sabatia and Burkhart 2014; Barrio-Anta et al. 2020).
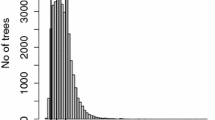
In this work, a Random Forest model was fitted using the R package randomForest (Liaw and Wiener 2002). For calibrating the number of trees, we fitted models with trees ranging from 100 to 10000 until we determined that 1000 trees were enough for the model results to be roughly stable, independently on the random seed. We calibrated the predictors’ subset size at each split, mtry, by trying different values ranging from 1/2 to 1/6 of the total amount of predictors. In order to enhance the stability of these fittings, we performed a repeated tenfold cross-validation. For selecting the least amount of necessary predictors, we carried out a recursive variable elimination (RVE) procedure similar to the one applied by Weiskittel et al. (2011). At each step of this procedure, the least important predictor (measured by the decrease in accuracy due to its removal from the model) was dropped off from the set. Then, a new model with the remaining predictors was fitted. This was repeated until there were only two predictors left. The best alternative along the RVE path was considered to be the one that provided a reasonably high predictive performance, subjected to have the least amount of predictors. The criteria we used for this purpose was to look for the model previous to a significant decrease in \(R^2\) (square of the Pearson’s correlation between observed and predicted SI) due to the removal of a certain predictor. Specifically, we selected the model for which dropping off one predictor produced a decrease in \(R^2\) higher than the 90th percentile of all the decreases distribution in the RVE path.
2.3.2 Boosted trees regression
Similarly to Random Forest, Boosted Trees (Valiant 1984) is a rule-based ensemble technique that combines multiple weak learners (regression trees) to enhance performance in the final prediction. Unlike Random Forest, Boosted Trees ensemble is hierarchized, being each additional tree a regressor of predictive residuals of previous trees.
In this study, we fitted a Boosted Trees model using the R package gbm (Greenwell et al. 2019). Firstly, we set the optimum values of the calibration constants through a 10 times repeated tenfold cross validation (maximising \(R^2\)). These constants were: number of iterations (i.e., number of trees), interaction depth (maximum number of levels of nested nodes in each tree), and shrinkage (penalization applied to each tree’s residual prediction). Once the calibration constants were defined, we performed an RVE identical to the one applied to Random Forest for selecting the minimum necessary number of predictors.
2.3.3 Cubist regression
Cubist (Quinlan 1992) is a rule-based technique that performs a boosting-like procedure for enhancing the predictive performance by nestedly re-predicting the residuals throughout a defined number of iterations usually known as “committees”. The significant difference with Boosted Trees is that in Cubist the fitted models for re-predicting residuals are not proper regression trees but a tree-like hierarchized ensemble of linear models. At each tree split, a linear model (which may not include the node predictor) is fitted for predicting the response. As the linear models at the branch ends of trees can make predictions outside the response observed range, a correction factor usually called extrapolation correction can be applied for controlling the coherency of the predicted values. For allowing this approach to be comparable to the Random Forest and Boosted Trees fitted models, we set the extrapolation correction as a fixed value of 100, which means that the predicted values will be strictly forced to be inside the observational range. Cubist algorithm also includes a nearest-neighbor parameter that allows for correcting each prediction based on similarity to observations used in the training set.
We fitted a Cubist model using the R package Cubist (Kuhn and Quinlan 2018). Firstly, we calibrated the number of committees and the number of neighbors for correction through a 10-times repeated 10-fold cross validation (based on \(R^2\) maximization). After this, we performed an RVE procedure similar to the ones applied to Random Forest and Boosted Trees. The importance of each predictor was measured by the usage rate, which indicates the frequency of each predictor as node-ruler or as linear explainer throughout the committees.
The RVE path of the three rule-based models fitted is shown in Fig. 3.
2.4 SI indirect prediction
We proposed an alternative indirect methodology for SI prediction based on multivariate linear models. This methodology consisted of two stages: (1) predicting the nine growth parameters we previously estimated (a, b, c, \(h_{ip}\), \(t_{ip}\), \(g_{ip}\), \(h_{mmg}\), \(t_{mmg}\), mmg) as a function of the WorldClim 2 bioclimatic variables, and (2) to estimate SI as a function of the new climate-sensitive predictions of growth parameters (\(\hat{a}\), \(\hat{b}\), \(\hat{c}\), \(\hat{h}_{ip}\), \(\hat{t}_{ip}\), \(\hat{g}_{ip}\), \(\hat{h}_{mmg}\), \(\hat{t}_{mmg}\), \(\hat{mmg}\)).
For the first stage, we used stepwise regression technique using the R language package stats (R Core Team 2018) for carrying out a generalized least squares coefficients estimation (generalized linear model, GLM) and automatic variable selection. This technique has been recurrently used in forestry, and specifically for SI-environment modeling (Codilan et al. 2015; Tange and Ge 2020). For attaining model parsimony, we used Akaike’s information criterion (AIC), so the fitting procedure involved the minimization of the following loss function:
being k a penalty p the number of parameters in the model, N the number of observations, and MSE the mean square error. For calibrating k, we used a tenfold cross-validation procedure. Within this procedure, we chose the best k for each case that maximized the \(R^2\), submitted to provide significant coefficients at a minimum confidence level of 90%.
Regarding the second stage of indirect SI modeling, we developed different transformations derived from Eq. (2) for extracting SI out of the predicted values of the nine alternative growth parameters. From Eq. (2), we can estimate the site index as:
being a, b and c the previously fitted parameters for each plot. Considering this, we could also perform an indirect climate-sensitive SI prediction (\(SI_{abc}\)) as follows:
being \(\hat{a}\), \(\hat{b}\) and \(\hat{c}\) the growth parameters predicted as a function of the bioclimatic explainers.
Combining Eq. (4) with the calculation methods summarized in Table 2 allowed us for predicting SI from different sets of three growth parameters. We accomplished this by isolating a and b parameters from equations in Table 2 and substituting in Eq. (4). As a result, we developed six different methods for estimating SI from the nine alternative growth parameters predicted, which are summarized in Table 4.
As a “control” method for allowing a reliable comparison between direct rule-based and indirect parametric models, we also fitted a direct stepwise regression (GLM) for predicting SI as a function of the 19 WorldClim predictors using the same procedure described in this section for predicting the nine growth parameters.
2.5 Model evaluation
For testing the robustness of the fitted models we performed a bootstrap validation procedure based on the 632+ rule (Efron and Tibshirani 1997), which we already tested in a previous study (González-Rodríguez and Diéguez-Aranda 2020). We calculated the bootstrap error of each model fitted through a resampling set of 100 realizations per model using the R package boot (Canty and Ripley 2017). Then, we estimated the overall predicitive error of each model by doing a weighted mean of statistics between apparent and bootstrap performance:
where MSE\(_{training}\) is the apparent mean square error (MSE), MSE\(_{bootstrap}\) is the mean bootstrap MSE, MSE\(_{632+}\) is the corrected MSE, and w is the weight parameter that accounts for the observed relative overfitting and the no information rate (i.e., the potential error if observed and predicted values were completely uncorrelated).
Once we carried out the bootstrap validation, we examined the plots of residuals to detect possible patterns of heteroscedasticity or regression to the mean in the fitted models. In addition, we assessed the role of each predictor within the SI model with the best validation performance in order to evaluate its ecological coherence. Considering the potential difficulty of directly understanding the behaviour of the fitted models, we based our interpretation on the visualization of standardized predictors against predicted SI values. To facilitate this task, we preferred to focus on LOESS fitted curves rather than directly analysing the point clouds. This interpretation method gave us an overview of the role of each predictor throughout the different site conditions that exist in the training dataset.
3 Results
A summary of goodness-of-fit statistics for the SI models fitted is shown in Table 5. Regarding model calibration, Random Forest had an optimum mtry value of 2/3 and Boosted Trees had optimum values of 20 for the number of iterations (trees), one for the interaction depth and 0.1 for the shrinkage penalization. The optimum constants for the Cubist model where 20 committees and nine neighbors for correction. The parameter estimates and p values of the stepwise models fitted for predicting the nine growth parameters are presented in the Appendix (Table 6). All the slope parameters estimated for these models were significant at least at 95% level of confidence. The average amount of predictors per models was three, having a maximum of seven for the \(t_{ip}\) model and a minimum of zero for the c model, which resulted a null model.
Concerning model performance, the rule-based approaches provided the highest apparent values of \(R^2\), having the Random Forest model the best performance (\(R^2\)=0.802), followed by Boosted Trees and Cubist. In contrast, the indirect parametric models had significantly smaller values of apparent \(R^2\), with a maximum value of 0.384 for the case of MMG1. The direct GLM had apparent performance slightly higher than MMG1 (\(R^2\)=0.414), but much lower than the rule-based approaches. Overall, the models fitted had a number of predictors ranging from five (MMG1, MMG2, MMG and Boosted Trees models) to nine (IP1, IP2 models). Considering our variable selection criterion, the three rule-based models fitted reached the maximum cross-validated performance using three predictors, which were, in all cases, BIO1, BIO2 and BIO3. The AIC-based variable selection for the direct GLM also produced an optimum number of predictors equal to three, which where BIO7, BIO8, BIO9.
Concerning bootstrap validation, rule-based models presented a very high relative overfitting rate, ranging from 54% to 72%. Among these models, Cubist resulted in the model with the best performance both in NRMSE\(_{632+}\) (0.206) and \(R^2_{632+}\) (0.285) and also with the lowest relative overfitting rate (54%). The direct GLM seemed extremely prone to overfitting, with a very low validation performance (\(R^2_{632+}\) = 0.136) and a relative overfitting rate reaching 80%. In comparison, the fitted indirect parametric models produced more variable bootstrap performances, with \(R^2_{632+}\) values ranging from 0.187 to 0.298, and relative overfitting rates (R) ranging from 0.311 to 0.581. Models ABC, MMG2, and MMG3 produced low performances but also had very high overfitting rates. Model IP2 had a poor apparent performance but also showed a low relative overfitting rate. Models MMG1 and IP1 were the ones with the best overall bootstrap performance, presenting a high \(R^2_{632+}\), a low NRMSE\(_{632+}\) and a moderate relative overfitting rate. Considering that the performance estimates of MMG1 were slighltly higher than IP2, we selected this model as the best parametric alternative for subsequent analyses. The final form of MMG1 model, after combining the corresponding equation in Table 4 with the linear models fitted was as follows:
where \(a_{0}\)=-117.677 , \(a_{1}\)= 5.132701, \(a_{2}\)=0.159120 , \(a_{3}\)=-8.09051 , \(a_{4}\)= 0.00283399, \(a_{5}\)=46.4528 , \(a_{6}\)=1.34853 , \(a_{7}\)=-47.092005 , and \(a_{8}\)= 1.69057.
Plots of observed SI vs predicted SI and predicted SI vs residuals for Cubist and MMG1 models are shown in Fig. 4. After analyzing the model residuals of both approaches, we did not found any significant trace of heteroscedasticity. In order to support model interpretation, a plot of LOESS curves that represent predicted SI vs. standardized predictors for MMG1 model is presented in Fig 5.
Plots of: a observed SI vs predicted SI for MMG1; b predicted SI vs residuals for MMG1; c observed SI vs predicted SI for Cubist; and d predicted SI vs residuals for Cubist. The dashed line in plots a and c represents the linear trend of observed vs predicted SI, while the solid line in plots b and d represents the LOESS-smoothed trend of residuals vs predicted SI
4 Discussion
The fitted models explained from 10% to 80% of the SI variability. The apparent performance range of the rule-based models fitted is similar to the observed in other studies for Random Forest (\(R^2\)=0.68, Weiskittel et al. 2011; \(R^2\)=0.59, Barrio-Anta et al. 2020) and Boosted Trees (\(R^2\) from 0.44 to 0.64, Aertsen et al. 2010). Similarly, the performance of the parametric models fitted (\(R^2\sim\) 0.27-0.38) was on average near the spectrum that can be found in literature (24%-27%, Monserud et al. 2006, 31%-52%, Aertsen et al. 2010, 34%-42%, Sabatia and Burkhart 2014 of explained variability).
Among the rule-based approaches, Random Forest showed the best apparent \(R^2\). However, its bootstrap validation performance was very similar to Boosted Trees and slightly lower than Cubist, which, despite being the model with the lowest apparent performance of the three, resulted in the most robust alternative. Though the differences in \(R^2_{632+}\) between the three tested techniques might be too slight to undoubtedly conclude that Cubist is the most suitable rule-based technique for SI-environment modeling, its lesser tendency to overfitting should be considered an important advantage. However, even considering the better robusticity shown by Cubist, the relative overfitting rates found for rule-based techniques are still very high in comparison with the ones provided by some indirect parametric models. We think that this could be an important concern for using these rule-based models for predicting SI out of the frame of this study. Using a complementary dataset for validation would be a necessary line of action for addressing this uncertainty in further research.
Regarding indirect parametric models, we observed that models based on the combination of growth + age (IP2 and MMG2 models) had lower performance. In contrast, combinations of height + age (IP1 and MMG1) had the highest performance. The fact that MMG1 outperformed the rest of the parametric models, both in apparent and validation statistics, may suggest that \(h_{mmg}\) and \(t_{mmg}\) are more effective than the rest of growth parameters for representing growth–climate relationships. In this context, using \(h_{mmg}\) and \(t_{mmg}\) directly for dominant height projection—as \(t_1\) and \(H_1\) inside Eq. 1—would be a reasonable modeling alternative to explore in further research.
Despite the higher apparent performance of the Random Forest model, the observed robustness of MMG1 may make this parametric approach a better alternative for SI prediction. This finding runs parallel to the results found by Sabatia and Burkhart (2014), where a parametric model outperformed Random Forest, in terms of robustness and reliability. After checking the observed vs predicted SI values (Fig. 4), we found a moderate regression to the mean in both models. Besides, the range of predicted values was slightly narrower for Random Forest than for the parametric approach. This regression to the mean was also found in previous studies (Hamel et al. 2004; Weiskittel et al. 2011), and it has been suggested to be a common characteristic of the SI-environment models (Sabatia and Burkhart 2014).
The comparison between performance statistics of the direct GLM and MMG1 revealed that the indirect approach was more robust, being less prone to overfitting than the direct model. We think this finding might imply that, indeed, the indirect methods could be more effective at capturing the existing relationships between tree growth and climate and, hence, more ecologically consistent than direct SI modeling approaches. However, the proposed indirect method has got also some potential drawbacks. Regarding data acquisition, stem analysis is much more expensive and technically complex than a simple forest inventory of temporal research plots. We think that this is a crucial aspect to consider for practical applications of indirect SI modeling.
Concerning the ecological interpretation of MMG1 model, assessing the role of each predictor in Eq. 7 is not a straightforward task, as some predictors appear both in the numerator and in the denominator of the model form, apparently with a similar behaviour. The analysis of LOESS curves in Fig. 5 revealed that, overall, BIO2 and BIO3 had a strong positive influence on SI. As these variables are proportional to thermal diurnal ranges, and therefore potential indicators of altitude and continentality (Oliver 2005), we propose two possible reasons for their impact on SI: (1) altitude can imply cooler winter and night temperatures, which is positive for satisfying the chilling needs of the species, and (2) continentality may imply the absence of salty sea winds, potentially harmful for Scots pine (Øyen et al. 2006; Savill 2013). A special comment about the chilling effect is needed. Admittedly, certain conifer species, especially those naturally distributed in cold regions, such as Scots pine (Øyen et al. 2006), might suffer from a certain stress on carbon balance due to high respiration rates during mild winters (Pâques 2013; Smith et al. 1995). We already found this to be an important restriction for growth of radiata pine in the same region in a previous study (González-Rodríguez and Diéguez-Aranda 2020). We think this may be a particular feature of the studied region, characterized by a very humid and temperate-warm oceanic climate (predominantly Csb climate with Cfa and Cfb local variations, according to the Köppen-Geiger classification, updated by Kottek et al. 2006).
The predictor BIO9 showed, overall, a very slight positive influence on SI, occurring mostly on the second half of its range. We hypothesize that this slight trend may represent the positive influence of temperatures for growth during the growing season, specially for coldest sites at higher altitudes (corresponding to Cfb local variants). Thought this predictor should also capture the negative effect of summer drought stress on growth, the high precipitation registered in the set of sampled sites—with a minimum of 1246 mm—may make the latter a not significant constraint for growth. BIO4 and BIO7 showed a similar influence on predicted SI, having a maximum at the middle of their ranges, showing, respectively, a positive and negative contribution on growth towards the extremes. In the case of BIO7, low values of this predictor may have a negative influence on SI because of the same reason explained for BIO2 and BIO3. High values of BIO7 may contribute negatively to SI representing the effect of frost stress factors in very contrasted temperature regimes. Finally, concerning BIO4, the positive effect on SI of its left tail may be also related to the influence of frost stress in sites with very regular precipitation regimes, associated with high altitudes (Cfb climates).
5 Conclusion
We fitted a set of rule-based and indirect parametric models for predicting site index (SI) of Scots pine stands as a function of bioclimatic variables. The models fitted explained from \(\sim\)10% to \(\sim\)80% of the response’s variability. The rule-based approaches tested showed very high apparent performance statistics, being Random Forest the one with the highest \(R^2\). However, the bootstrap validation procedure carried out revealed that these techniques were also very prone to overfitting, and besides their actual differences in performance were little. In contrast, indirect parametric models showed a much lower overfitting rate. Two of these models, MMG1 and IP1, had better bootstrap error estimates than the rule-based approaches. Specifically, MMG1, a parametric model derived from height and age at the maximum mean growth point, showed the highest validation \(R^2\) among the set of fitted models, explaining up to 38% of the SI variability. According to this model, SI was mainly conditioned by different measures of continentality, but also by heat and rainfall variables. We concluded that, for the specific scope of our study, the use of an indirect approach based on easily interpretable parametric models was a better modeling alternative than the direct prediction of SI using rule-based techniques.
Data availability
The datasets analysed during the current study are available in the Zenodo repository, https://doi.org/10.5281/zenodo.4535243.
References
Aertsen W, Kint V, van Orshoven J, Özkan K, Muys B (2010) Comparison and ranking of different modelling techniques for prediction of site index in Mediterranean mountain forests. Ecol Model. https://doi.org/10.1016/j.ecolmodel.2010.01.007
Barrio-Anta M, Castedo-Dorado F, Cámara-Obregón A, López-Sánchez CA (2020) Predicting current and future suitable habitat and productivity for Atlantic populations of maritime pine (Pinus pinaster Aiton) in Spain. Ann For Sci. 77(2):41. https://doi.org/10.1007/s13595-020-00941-5, https://link.springer.com/10.1007/s13595-020-00941-5
Bontemps JD, Bouriaud O (2014) Predictive approaches to forest site productivity: Recent trends, challenges and future perspectives. Forestry 87(1):109–128. https://doi.org/10.1023/A:1010933404324
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://link.springer.com/article/10.1023/A:1010933404324, https://doi.org/10.1023/A:1010933404324
Canty A, Ripley BD (2017) boot: Bootstrap R (S-Plus) Functions
Carmean W (1972) Site Index Curves for Upland Oaks in the Central States. For Sci. https://doi.org/10.1093/forestscience/18.2.109
Codilan AL, Nakjima T, Tatsuhara S, Shiraisi N (2015) Estimating site index from ecological factors for industrial tree plantation species in Mindanao, Philippines. Bull. Univ. of Tokyo For 133:19–41
Crookston NL, Rehfeldt GE, Dixon GE, Weiskittel AR (2010) Addressing climate change in the forest vegetation simulator to assess impacts on landscape forest dynamics. For Ecol Manag 260(7):1198–1211. https://doi.org/10.1016/j.foreco.2010.07.013
Diéguez-Aranda U, Álvarez González JG, Barrio Anta M, Rojo Alboreca A (2005a) Site quality equations for Pinus sylvestris L. plantations in Galicia (northwestern Spain). Ann For Sci. https://doi.org/10.1051/forest:2005006
Diéguez-Aranda U, Burkhart HE, Rodríguez-Soalleiro R (2005b) Modeling dominant height growth of radiata pine (Pinus radiata D. Don) plantations in north-western Spain. For Ecol Manag. https://doi.org/10.1016/j.foreco.2005.05.015
Diéguez-Aranda U, Castedo Dorado F, Álvarez González JG, Rojo Alboreca A (2006) Dynamic growth model for Scots pine (Pinus sylvestris L.) plantations in Galicia (north-western Spain). Ecol Model. https://doi.org/10.1016/j.ecolmodel.2005.04.026
Efron B, Tibshirani R (1997) Improvements on cross-validation: The .632+ bootstrap method. J Am Stat Assoc. https://doi.org/10.1080/01621459.1997.10474007, arXiv:1011.1669v3
Fick SE, Hijmans RJ (2017) WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. https://doi.org/10.1002/joc.5086
Fontes L, Bontemps JD, Bugmann H, Van Oijen M, Gracia C, Kramer K, Lindner M, Rotzer T, Skovsgaard JP (2010) Models for supporting forest management in a changing environment. Forest Systems 19:8–29. https://doi.org/10.5424/fs/201019S-9315
Gompertz B (1825) On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. Phil Transac Roy Soci Londo 115:513–585. https://doi.org/10.1098/rspl.1815.0271
González-Rodríguez M, Diéguez-Aranda U (2020) Exploring the use of learning techniques for relating the site index of radiata pine stands with climate, soil and physiography. For Ecol Manag. p 458. https://doi.org/10.1016/j.foreco.2019.117803
González-Rodríguez M, Dieguez-Aranda U (2021) Height growth paramters of Scots pine plantations in the north-west of Spain from plot measurements and stem analysis [Data set]. https://doi.org/10.5281/zenodo.4535243
Greenwell B, Boehmke B, Cunningham J, Developers GBM (2019) gbm: Generalized Boosted Regression Models. https://cran.r-project.org/package=gbm
Hamel B, Bélanger N, Paré D (2004) Productivity of black spruce and Jack pine stands in Quebec as related to climate, site biological features and soil properties. For Ecol Manag. https://doi.org/10.1016/j.foreco.2003.12.004
Hossfeld J (1822) Mathematika für Forstmänner, Ö konomen und Cameralisten
Kirilenko AP, Sedjo RA (2007) Climate change impacts on forestry. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.0701424104
Kottek M, Grieser J, Beck C, Rudolf B, Rubel F (2006) World Map of Köppen-Geiger Climate Classification - (updated with CRU TS 2.1 temperature and VASClimO v1.1 precipitation data 1951 to 2000). Meteorologische Zeitschrift
Kuhn M, Quinlan R (2018) Cubist: Rule- And Instance-Based Regression Modeling. https://cran.r-project.org/package=Cubist
Liaw A, Wiener M (2002) Classification and Regression by randomForest. R News 2(3):18–22. https://cran.r-project.org/doc/Rnews/
Lindner M, Garcia-Gonzalo J, Kolström M, Green T, Reguera R, Maroschek M, Seidl R, Lexer MJ, Netherer S, Schopf A, Kremer A, Delzon S, Barbati A, Marchetti M, Corona P (2008) Impacts of climate change on european forests and options for adaptation. Report to the European Commission Directorate-General for Agriculture and Rural Development
Monserud RA, Huang S, Yang Y (2006) Predicting lodgepole pine site index from climatic parameters in Alberta. For Chron 82(4):562–571. https://doi.org/10.5558/tfc82562-4
Newberry JD (1991) A note on Carmean’s estimate of height from stem analysis data. https://doi.org/10.1093/forestscience/37.1.368
Oliver JE (ed) (2005) Encyclopedia of world Climatology, 1st edn. Springer, Netherlands, Dordrecht
Øyen BH, Blom HH, Gjerde I, Myking T, Sætersdal M, Thunes KH (2006) Ecology, history and silviculture of Scots pine (Pinus sylvestris L.) in western Norway - A literature review. Forestry 79(3):319–329. https://doi.org/10.1093/forestry/cpl019
Pâques LE (2013) Forest tree breeding in Europe: Current state-of-the-art and perspectives. https://doi.org/10.1007/978-94-007-6146-9\_9
Quinlan JR (1992) Learning with continuous classes. Fifth Austrailian Joint Conference of Artifical Intelligence 92:343–348
R Core Team (2018) R: A Language and Environment for Statistical Computing. https://www.r-project.org/
Richards FJ (1959) A flexible growth function for empirical use. J Exp Bot. https://doi.org/10.1093/jxb/10.2.290
Sabatia CO, Burkhart HE (2014) Predicting site index of plantation loblolly pine from biophysical variables. For Ecol Manag. 326:142–156. https://doi.org/10.1016/j.foreco.2014.04.019
Savill PS (2013) The silviculture of trees used in British forestry, 2nd edn. CABI Publishing, Oxfordshire
Seynave I, Gégout JC, Hervé JC, Dhôte JF, Drapier J, Bruno É, Dumé G (2005) Picea abies site index prediction by environmental factors and understorey vegetation: a two-scale approach based on survey databases. Can J For Res 35(7):1669–1678. https://doi.org/10.1139/x05-088
Skovsgaard JP, Vanclay JK (2008) Forest site productivity: A review of the evolution of dendrometric concepts for even-aged stands. Forestry 81(1):13–31. https://doi.org/10.1093/forestry/cpm041
Smith WK, Roy J, Hinckley TM (1995) Ecophysiology of Coniferous Forests. Elsevier, https://doi.org/10.1016/C2009-0-02453-2, https://linkinghub.elsevier.com/retrieve/pii/C20090024532
Swenson JJ, Waring RH, Fan W, Coops N (2005) Predicting site index with a physiologically based growth model across Oregon, USA. Can J For Res. 35(7):1697–1707. https://doi.org/10.1139/x05-089, http://www.nrcresearchpress.com/doi/10.1139/x05-089
Tange T, Ge F (2020) Topographic factors and tree heights of aged Cryptomeria japonica plantations in the Boso Peninsula, Japan. Forests 11(7). https://doi.org/10.3390/F11070771
Valiant LG (1984) A theory of the learnable. In: Proceedings of the sixteenth annual ACM symposium on Theory of computing - STOC ’84. https://doi.org/10.1145/800057.808710
Wang GG, Huang S, Monserud RA, Klos RJ (2004) Lodgepole pine site index in relation to synoptic measures of climate, soil moisture and soil nutrients. For Chron 80(6):678–686. https://doi.org/10.5558/tfc80678-6
Watt MS, Dash JP, Bhandari S, Watt P (2015) Comparing parametric and non-parametric methods of predicting Site Index for radiata pine using combinations of data derived from environmental surfaces, satellite imagery and airborne laser scanning. For Ecol Manag. https://doi.org/10.1016/j.foreco.2015.08.001
Watt MS, Dash JP, Watt P, Bhandari S (2016) Multi-sensor modelling of a forest productivity index for radiata pine plantations. N Z J For Sci. 46(1). https://doi.org/10.1186/s40490-016-0065-z
Watt MS, Palmer DJ, Leonardo EMC, Bombrun M (2021) Use of advanced modelling methods to estimate radiata pine productivity indices. For Ecol Manag. 479:118557. https://doi.org/10.1016/j.foreco.2020.118557, https://linkinghub.elsevier.com/retrieve/pii/S0378112720313268
Weiskittel AR, Crookston NL, Radtke PJ (2011) Linking climate, gross primary productivity, and site index across forests of the western United States. Can J For Res 41(8):1710–1721. https://doi.org/10.1139/x11-086, http://www.nrcresearchpress.com/doi/abs/10.1139/x11-086
Zhu G, Hu S, Chhin S, Zhang X, He P (2019) Modelling site index of Chinese fir plantations using a random effects model across regional site types in Hunan province, China. For Ecol Manag 446:143–150. https://doi.org/10.1016/j.foreco.2019.05.039
Acknowledgements
We thank the authors of Worldclim 2 climate dataset (Fick and Hijmans 2017), for the bioclimatic maps provided.
Funding
Plot data collection was funded by the Spanish Ministry of Science and Technology through the project AGL20013871-C02-01. The work of the first author and main researcher of this study has been partially funded by the Spanish Ministry of Industry, Economy and Competitiveness (DI-16-08971) and by the forest management consultancy company CERNA Ingenieri y Asesori Medioambiental S.L.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: John M Lhotka
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contribution of the co-authors Conceptualization: González-Rodríguez M.A., Diéguez-Aranda U.;
Methodology: González-Rodríguez M.A.;
Software: González-Rodríguez M.A.;
Validation: González-Rodríguez M.A.;
Software: González-Rodríguez M.A.;
Formal Analysis: González-Rodríguez M.A.;
Investigation: González-Rodríguez M.A.;
Resources: Diéguez-Aranda U.;
Data curation: González-Rodríguez M.A.;
Writing – original draft: González-Rodríguez M.A.;
Writing – review & editing: González-Rodrígue M.A., Diéguez-Aranda U.;
Visualisation: González-Rodríguez M.A.;
Supervision: Diéguez-Aranda U.;
Project Administration: Diéguez-Aranda U.;
Funding acquisition: Diéguez-Aranda U.
Appendix
Appendix
Rights and permissions
About this article
Cite this article
González-Rodríguez, M.Á., Diéguez-Aranda, U. Rule-based vs parametric approaches for developing climate-sensitive site index models: a case study for Scots pine stands in northwestern Spain. Annals of Forest Science 78, 23 (2021). https://doi.org/10.1007/s13595-021-01047-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13595-021-01047-2