Abstract
The validation of many anuran species is based on a strictly descriptive, morphological analysis of a small number of specimens with a limited geographic distribution. The Scinax Wagler, 1830 genus is a controversial group with many doubtful taxa and taxonomic uncertainties, due a high number of cryptic species. One example is the pair of species Scinax constrictus and Scinax nebulosus, which share a similar morphology. Scinax constrictus is restricted to the Brazilian Cerrado savanna, while S. nebulosus is widely distributed throughout northern South America. Despite the validation of many anuran species, discriminations based only on morphological traits is quite difficult due to the high conservative morphology of some groups. In this context, the present study uses mitochondrial and nuclear genes to provide a more consistent diagnosis and test the validity of S. constrictus as a distinct species from S. nebulosus, as well as evaluate the position of these taxa within the Scinax genus. The topologies obtained herein uphold the monophyletic status of Scinax based on all molecular markers assessed in this study, in all analytical approaches, with high levels of statistical support.
Keywords:
anurans; cryptic species; molecular markers; phylogeny; Scinax
Resumo
A validação de muitas espécies de anuros é baseada em uma análise morfológica e descritiva de um pequeno número de espécimes com uma distribuição geográfica limitada. O gênero Scinax Wagler, 1830 é um grupo controverso com muitos táxons duvidosos e incertezas taxonômicas devido ao grande número de espécies crípticas. Um exemplo são as espécies, Scinax constrictus e Scinax nebulosus, que compartilham uma morfologia similar. Scinax constrictus é restrito à savana do Cerrado brasileiro, enquanto S. nebulosus é amplamente distribuído pelo norte da América do Sul. Apesar da validação de muitas espécies de anuros, a discriminação baseada apenas em características morfológicas é bastante difícil, devido à alta morfologia conservadora de alguns grupos. Neste contexto, o presente estudo utiliza genes mitocondriais e nucleares para fornecer um diagnóstico mais consistente e para testar a validade de S. constrictus como uma espécie distinta de S. nebulosus, bem como avaliar a posição destes táxons dentro do gênero Scinax. As topologias obtidas confirmaram o status monofilético de Scinax com base em todos os marcadores moleculares, em todas as abordagens analíticas, com altos níveis de suporte estatístico.
Palavras-chave:
anuros; espécies crípticas; marcadores moleculares; filogenia; Scinax
1. Introduction
Phylogenetic systematics seeks evidence on the evolutionary relationships among species in order to comprehend diversity patterns (Lourenço et al., 2015LOURENÇO, L.B., TARGUETA, C.P., BALDO, D., NASCIMENTO, J., GARCIA, P.C., ANDRADE, G.V., HADDAD, C.F. and RECCO-PIMENTEL, S.M., 2015. Phylogeny of frogs the genus Physalaemus (Anura, Leptodactylidae) inferred from mitochondrial and nuclear gene sequences. Molecular Phylogenetics and Evolution, vol. 92, pp. 204-216. http://dx.doi.org/10.1016/j.ympev.2015.06.011. PMid:26143292.
http://dx.doi.org/10.1016/j.ympev.2015.0...
). Neotropical amphibians are characterized by considerable phylogeographic structuring and profound levels of genetic divergence, which require extended periods of time for the evolution of phenotypical differences (Sá et al., 2014SÁ, R.O., GRANT, T., CAMARGO, A., HEYER, R.W., PONSSA, M.L. and STANLEY, E., 2014. Systematics of the Neotropical Genus Leptodactylus Fitzinger, 1826 (Anura: Leptodactylidae): phylogeny, the relevance of non-molecular evidence, and species accounts. São Paulo: Sociedade Brasileira de Herpetologia.; Suk et al., 2020SUK, H.Y., BAE, H.G., KIM, D.Y., WON, H., BAEK, H.J., LEE, C.H., KIM, D.Y., GO, Y.M., SONG, J.Y., LEE, H. and MIN, M.S., 2020. Genetic and phylogenetic structure of Hynobius quelpaertensis, an endangered endemic salamander species on the Korean Peninsula. Genes & Genomics, vol. 42, no. 2, pp. 165-178. PMid:31797315.).
As a group, Neotropical amphibians present relatively conserved morphological characteristics and, in many cases, reliable diagnostic traits are difficult to identify (Nogueira et al., 2016NOGUEIRA, L., SOLÉ, M., SIQUEIRA, S., AFFONSO, P.R.A.M., STRÜSSMANN, C. and SAMPAIO, I., 2016. Genetic analysis reveals candidate species in the Scinax catharinae clade (Amphibia: Anura) from Central Brazil. Genetics and Molecular Biology, vol. 39, no. 1, pp. 49-53. http://dx.doi.org/10.1590/1678-4685-GMB-2015-0037. PMid:27007898.
http://dx.doi.org/10.1590/1678-4685-GMB-...
), which often implies the existence of cryptic species, which have been increasingly identified in recent years (Guarnizo et al., 2015GUARNIZO, C.E., PAZ, A., MUNOZ-ORTIZ, A., FLECHAS, S.V., MENDEZ-NARVAEZ, J. and CRAWFORD, A.J., 2015. DNA Barcoding Survey of Anurans across the Eastern Cordillera of Colombia and the Impact of the Andes on Cryptic Diversity. PLoS One, vol. 10, no. 5, pp. e0127312. http://dx.doi.org/10.1371/journal.pone.0127312. PMid:26000447.
http://dx.doi.org/10.1371/journal.pone.0...
; Pinto et al., 2019PINTO, B.J., COLLI, G.R., HIGHAM, T.E., RUSSELL, A.P., SCANTLEBURY, D.P., VITT, L.J. and GAMBLE, T., 2019. Population genetic structure and species delimitation of a widespread, Neotropical dwarf gecko. Molecular Phylogenetics and Evolution, vol. 133, pp. 54-66. http://dx.doi.org/10.1016/j.ympev.2018.12.029. PMid:30590108.
http://dx.doi.org/10.1016/j.ympev.2018.1...
). This means that the species-level diversity of many genera has long been underestimated. In this context, molecular analyses can be extremely useful for the identification of cryptic variations in morphologically identical populations (Lima, 2011LIMA, M.G., 2011. Variação geográfica de Scinax argyreornatus (Miranda- Ribeiro, 1926) (Anura: Hylidae). Recife: Universidade Federal de Pernambuco, 98 p. Dissertação de Mestrado em Biologia Animal.; Takazone, 2015TAKAZONE, A.M.G., 2015. Estudo taxonômico de Scinax nebulosus (SPIX, 1824) e Scinax constrictus (LIMA, BASTOS E GIARETTA, 2004) (ANURA, HYLIDAE) com base em parâmetros morfológicos, acústicos e de distribuição geográfica. São Luís: Universidade Federal do Maranhão, 77 p. Dissertação de Mestrado em Biodiversidade e Conservação.; Bessa-Silva et al., 2016BESSA-SILVA, A.R., VALLINOTO, M., SODRÉ, D., CUNHA, D.B., ASP, N.E., SAMPAIO, I., SCHNEIDER, H. and SEQUEIRA, F., 2016. Patterns of genetic variability in Island Populations of the Cane Toad (Rhinella marina) from the Mouth of the Amazon. PLoS One, vol. 11, no. 4, pp. e0152492. http://dx.doi.org/10.1371/journal.pone.0152492. PMid:27073849.
http://dx.doi.org/10.1371/journal.pone.0...
).
The Hylidae family is one of the most diverse anuran groups, found in Europe, northern Africa and Asia, South and Central America, western India, Australia, and New Guinea (Salducci et al., 2002SALDUCCI, M., MARTY, C., CHAPPAZ, R. and GILLES, A., 2002. Molecular phylogeny of French Guiana Hylinae: implications for the systematic and biodiversity of the Neotropical frogs. Comptes Rendus Biologies, vol. 325, no. 2, pp. 141-153. http://dx.doi.org/10.1016/S1631-0691(02)01423-3. PMid:11980175.
http://dx.doi.org/10.1016/S1631-0691(02)...
). The Hylidae group is currently one of the most diverse anuran families, comprising about 701 species, around 14.5% of all anurans. Almost half of these species (338 species in 27 genera) are found in Brazil (Frost, 2017FROST, D.R., 2017 [viewed 12 November 2017]. Amphibians species of the world 3.0: an online reference [online]. New York: American Museu of Natural History. Available from: http://research.amnh.org/herpetology/amphibia/index.php
http://research.amnh.org/herpetology/amp...
).
In the recently proposed arrangement by Duellman et al. (2016)DUELLMAN, W.E., MARION, A.B. and HEDGES, S.B., 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa, vol. 4104, no. 1, pp. 1-109. http://dx.doi.org/10.11646/zootaxa.4104.1.1. PMid:27394762.
http://dx.doi.org/10.11646/zootaxa.4104....
, the Scinax (Wagler, 1830) genus was excluded from the Hylinae subfamily and assigned to a new subfamily, Scinaxinae, which contains 70 described species (Frost, 2017FROST, D.R., 2017 [viewed 12 November 2017]. Amphibians species of the world 3.0: an online reference [online]. New York: American Museu of Natural History. Available from: http://research.amnh.org/herpetology/amphibia/index.php
http://research.amnh.org/herpetology/amp...
). This analysis also supported a new arrangement for species belonging to the Scinax genus, which were originally categorized into two groups, the Scinax ruber and Scinax catharinae clades. In this new classification, the entire catharinae clade was reclassified into the Ololygon genus (Duellman et al., 2016DUELLMAN, W.E., MARION, A.B. and HEDGES, S.B., 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa, vol. 4104, no. 1, pp. 1-109. http://dx.doi.org/10.11646/zootaxa.4104.1.1. PMid:27394762.
http://dx.doi.org/10.11646/zootaxa.4104....
).
Despite the scope of the review by Duellman et al. (2016)DUELLMAN, W.E., MARION, A.B. and HEDGES, S.B., 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa, vol. 4104, no. 1, pp. 1-109. http://dx.doi.org/10.11646/zootaxa.4104.1.1. PMid:27394762.
http://dx.doi.org/10.11646/zootaxa.4104....
, the validity of some taxa and their position in the Scinax genus remain unresolved. Scinax nebulosus Spix, 1824 and Scinax constrictus Lima et al., 2004 are part of the rostratus group, as well as Scinax rostratus Peters, 1863, Scinax boulengeri Cope, 1887, Scinax garbei Miranda-Ribeiro, 1926, Scinax kennedyi Pyburn, 1973, Scinax jolyi Lescure & Marty, 2000, Scinax pedromedinae Henle, 1991, Scinax proboscideus Brongersma, 1933, and Scinax sugillatus Duellman, 1973. Scinax nebulosus ranges from southeastern Venezuela, through the Guianas and lower Amazon basin to the state of Alagoas, in northeastern Brazil, with some occurrence records for the Bolivian Amazon (Faivovich et al., 2005FAIVOVICH, J., HADDAD, C.F.B., GARCIA, P.C.A., FROST, D.R., CAMPBELL, J.A. and WHEELER, W.C., 2005. Systematic review of the frog family Hylidae, with special reference to Hylinae: phylogenetic analysis and taxonomic revision. Bulletin of the American Museum of Natural History, vol. 294, no. 1, pp. 1-240. http://dx.doi.org/10.1206/0003-0090(2005)294[0001:SROTFF]2.0.CO;2.
http://dx.doi.org/10.1206/0003-0090(2005...
). Scinax constrictus is found exclusively in Cerrado savanna areas, and is amply distributed in the Brazilian state of Goiás.
The diagnostic characters traditionally used to identify S. nebulosus and S. constrictus are inadequate for the reliable differentiation of these species, leading to the uncertain taxonomic status of some specimens from populations historically identified as belonging to one of the two species. The increasing use of molecular data has reinforced the often enigmatic nature of amphibian morphology evolution (Menezes et al., 2016MENEZES, L., CANEDO, C., BATALHA-FILHO, H., GARDA, A.A., GEHARA, M. and NAPOLI, M.F., 2016. Multilocus phylogeography of the treefrog Scinax eurydice (Anura, Hylidae) reveals a plio-pleistocene diversification in the Atlantic Forest. PLoS One, vol. 11, no. 6, pp. e0154626. http://dx.doi.org/10.1371/journal.pone.0154626. PMid:27248688.
http://dx.doi.org/10.1371/journal.pone.0...
; Ferrão et al., 2016FERRÃO, M., COLATRELI, O., FRAGA, R., KAEFER, I., MORAVEC, J. and LIMA, A.L., 2016. High Species Richness of Scinax Treefrogs (Hylidae) in a Threatened Amazonian Landscape Revealed by an Integrative Approach. PLoS One, vol. 11, no. 11, pp. e0165679. http://dx.doi.org/10.1371/journal.pone.0165679. PMid:27806089.
http://dx.doi.org/10.1371/journal.pone.0...
), and has revitalized the study of amphibian taxonomy. Many groups of amphibians are morphologically conservative and this, alongside their high convergence degree, has resulted in numerous erroneous interpretations of anuran phylogeny when based solely on morphological characteristics (Bossuyt and Milinkovitch, 2000BOSSUYT, F. and MILINKOVITCH, M.C., 2000. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proceedings of the National Academy of Sciences of the United States of America, vol. 97, no. 12, pp. 6585-6590. http://dx.doi.org/10.1073/pnas.97.12.6585. PMid:10841558.
http://dx.doi.org/10.1073/pnas.97.12.658...
; Guarnizo et al., 2015GUARNIZO, C.E., PAZ, A., MUNOZ-ORTIZ, A., FLECHAS, S.V., MENDEZ-NARVAEZ, J. and CRAWFORD, A.J., 2015. DNA Barcoding Survey of Anurans across the Eastern Cordillera of Colombia and the Impact of the Andes on Cryptic Diversity. PLoS One, vol. 10, no. 5, pp. e0127312. http://dx.doi.org/10.1371/journal.pone.0127312. PMid:26000447.
http://dx.doi.org/10.1371/journal.pone.0...
).
As morphological characters are not adequate to reliably distinguish these species, the genetic validity of populations diagnosed as S. constrictus or S. nebulosus was tested in the present study using both nuclear and mitochondrial data, as the lack of genetic data for these species reinforces the need for molecular analyses, allowing for assessments concerning the species arrangement within the Scinax genus.
2. Material and Methods
2.1. Sample collection and molecular procedures
S. nebulosus and S. constrictus specimens were obtained through donations and exchanges with scientific institutions, and new specimens were also sampled in the field (Figure 1, Table 1). Their collection was authorized by the Chico Mendes Institute for Biodiversity Conservation (ICMBio), through permanent license number 52593–3. The sampling covers a wide range of localities within the geographic distribution of the species (Figure 1). The samples were complemented by Scinax sequences available in GenBank. It is important to note that this study does not aim to generate threats of extinction.
Sampling localities of the S. nebulosus and S. constrictus specimens included in the phylogenetic analyses.
In the case of the newly collected specimens, muscle tissue samples were extracted from the posterior members and conserved in absolute alcohol in 1.5 mL microtubes. The specimens themselves were fixed in 10% formalin and stored in 70% ethanol and subsequently catalogued and deposited at the Federal University of Maranhão (UFMA) scientific collection.
Pseudis paradoxa, representing a closely-related hylid genus, was used as the outgroup, based on the recent phylogenies published for the genus (Faivovich et al., 2005FAIVOVICH, J., HADDAD, C.F.B., GARCIA, P.C.A., FROST, D.R., CAMPBELL, J.A. and WHEELER, W.C., 2005. Systematic review of the frog family Hylidae, with special reference to Hylinae: phylogenetic analysis and taxonomic revision. Bulletin of the American Museum of Natural History, vol. 294, no. 1, pp. 1-240. http://dx.doi.org/10.1206/0003-0090(2005)294[0001:SROTFF]2.0.CO;2.
http://dx.doi.org/10.1206/0003-0090(2005...
; Pyron and Wiens, 2011PYRON, R.A. and WIENS, J.J., 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Molecular Phylogenetics and Evolution, vol. 61, no. 2, pp. 543-583. http://dx.doi.org/10.1016/j.ympev.2011.06.012. PMid:21723399.
http://dx.doi.org/10.1016/j.ympev.2011.0...
; Duellman et al., 2016DUELLMAN, W.E., MARION, A.B. and HEDGES, S.B., 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa, vol. 4104, no. 1, pp. 1-109. http://dx.doi.org/10.11646/zootaxa.4104.1.1. PMid:27394762.
http://dx.doi.org/10.11646/zootaxa.4104....
). Genomic DNA was isolated from muscle tissue using a phenol-chloroform protocol modified from Sambrook and Russel (2001)SAMBROOK, J. and RUSSEL, D.W., 2001. Rapid isolation of yeast DNA. In: J. SAMBROOK and D.W. RUSSEL, eds. Molecular cloning, a laboratory manual. New York: Cold Spring Harbor Laboratory, pp. 631-632.. As negative amplification of some samples was noted, a Wizard® Genomic DNA Purification kit was applied, using the mouse tail protocol. DNA extraction confirmation and the quantification and quality determination of the extracted material was based on electrophoresis runs in 1% agarose gels.
2.2. Amplification and sequencing
Two mitochondrial genes (16S rRNA and Cytochrome Oxidase I (COI)) and the nuclear gene Rhodopsin were analyzed. Each gene was amplified with a specific pair of primers. The primers described by Palumbi et al. (1991)PALUMBI, S., MARTIN, A., ROMANO, S., MACMILLAN, W.O., STICE, L. and GRABOWSKI, G., 1991. The simple fool’s guide to PCR, Version 2.0. Honolulu: Department of Zoology and Kewalo Marine Laboratoly, University of Hawaii. were used for the 16S region, namelu L1987–5’ GCCTCGCCTGTTTACCAAAAAC 3’ and H2609–5’-CCGGTCTGAACTCAGATCACGT 3’. In the case of COI, the primers LCO-1490 5’GGTCAACAAATCATAAAGATATTGG 3’ and HCO-2198 5’TAAACTTCAGGGTGACCAAAAAATCA 3’ were used (Folmer et al., 1994FOLMER, O., BLACK, M., HOEH, W., LUTZ, R. and VRIJENHOEK, R., 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, vol. 3, no. 5, pp. 294-299. PMid:7881515.), and in the case of Rhodopsin, the primers consisted of Rhod1A 5’ACCATGAACGGAACAGAAGGYCC 3’ and Rhod1C 5’ CCAAGGGTAGCGAAGAARCCTTC 3’, described by Bossuyt and Milinkovitch (2000)BOSSUYT, F. and MILINKOVITCH, M.C., 2000. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proceedings of the National Academy of Sciences of the United States of America, vol. 97, no. 12, pp. 6585-6590. http://dx.doi.org/10.1073/pnas.97.12.6585. PMid:10841558.
http://dx.doi.org/10.1073/pnas.97.12.658...
.
The sequencing reactions were prepared using the ABI Dye Terminator kit (Applied Biosystems), following the manufacturer’s recommendations. Once amplified, the samples were injected into an ABI 3500XL automatic sequencer, resulting in a file with base sequences and the chromatogram.
It is important to note that, while an attempt was made to extract genomic DNA from all the samples and amplify all three markers, in some cases specimen conditions or tissue storage altered the quality of the extracted DNA, which was reflected in the amplification resolution.
2.3. Phylogenetic analyses
The DNA sequences were visualized using the Codoncode 3.0 (Codoncode Corporation), and aligned in MAFFT (Katoh and Standley, 2013KATOH, K. and STANDLEY, D.M., 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Molecular Biology and Evolution, vol. 30, no. 4, pp. 772-780. http://dx.doi.org/10.1093/molbev/mst010. PMid:23329690.
http://dx.doi.org/10.1093/molbev/mst010...
), followed by visual inspection to detect possible base identification errors. Variable sites were checked using the MEGA 7.0 software (Kumar et al., 2016KUMAR, S., STECHER, G. and TAMURA, K., 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution, vol. 33, no. 7, pp. 1870-1874. http://dx.doi.org/10.1093/molbev/msw054. PMid:27004904.
http://dx.doi.org/10.1093/molbev/msw054...
), and genetic distance values were also calculated using this program
Two different phylogenetic inferences were applied herein, namely Bayesian Inferences (BI), performed using the Mr. Bayes v.3.2 software (Ronquist et al., 2012RONQUIST, F., TESLENKO, M., VAN DER MARK, P., AYRES, D.L., DARLING, A., HÖHNA, S., LARGET, B., LIU, L., SUCHARD, M.A. and HUELSENBECK, J.P., 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, vol. 61, no. 3, pp. 539-542. http://dx.doi.org/10.1093/sysbio/sys029. PMid:22357727.
http://dx.doi.org/10.1093/sysbio/sys029...
) and a Maximum Likelihood (ML) analysis, using the PhyML v.3.0 program (Guindon et al., 2010GUINDON, S.J.F., DUFAYARD, V., LEFORT, M., ANISIMOVA, W., HORDIJK, W. and GASCUEL, O., 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, vol. 59, no. 3, pp. 307-321. http://dx.doi.org/10.1093/sysbio/syq010. PMid:20525638.
http://dx.doi.org/10.1093/sysbio/syq010...
). The most appropriate evolutionary model for each database was identified using jModeltest 2 (Darriba et al., 2012DARRIBA, D., TABOADA, G.L., DOALLO, R. and POSADA, D., 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, vol. 9, no. 8, pp. 772. http://dx.doi.org/10.1038/nmeth.2109. PMid:22847109.
http://dx.doi.org/10.1038/nmeth.2109...
). For the ML analysis, the reliability of each node was estimated using a nonparametric bootstrap procedure comprising 1000 replicates (Felsenstein, 1985FELSENSTEIN, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution; International Journal of Organic Evolution, vol. 39, no. 4, pp. 783-791. http://dx.doi.org/10.1111/j.1558-5646.1985.tb00420.x. PMid:28561359.
http://dx.doi.org/10.1111/j.1558-5646.19...
). The Bayesian Inferences were based on MCMC sampling in four simultaneous runs, each with four chains (one hot and three heated) with a total run of 10 million generations. The Bayesian probabilities were selected based on a consensus of 60%. As a conservative measure, 25% of the initial trees were discarded as burn-in. The log-likelihood scores were plotted in the Tracer v.1.5 program (Rambaut and Drummond, 2009RAMBAUT, A. and DRUMMOND, A.J., 2009 [viewed 12 November 2017]. Tracer version 1.5 [software]. Available from: http://beast.bio.ed.ac.uk
http://beast.bio.ed.ac.uk...
) to confirm the validity of the initial burn-in period used in Mr. Bayes. The post burn-in samples were used to build the trees used in the present study.
As the number of sequences obtained for the different markers varied, the results were presented in two different ways: (i) mitochondrial gene trees and (ii) a tree combining the 16S and Rhodopsin genes. In the latter case, only S. constrictus and S. nebulosus specimens in which both the 16S and Rhodopsin genes were amplified were included in the analyses.
Inconsistencies among gene trees and in species analyses based on multiple loci are relatively common in evolutionary studies (Nichols, 2001NICHOLS, R., 2001. Gene trees and species trees are not the same. Trends in Ecology & Evolution, vol. 16, no. 7, pp. 358-364. http://dx.doi.org/10.1016/S0169-5347(01)02203-0. PMid:11403868.
http://dx.doi.org/10.1016/S0169-5347(01)...
). In the present study, the phylogenetic reconstructions were compared using the BEAST v. 1.6.2 software (Rambaut and Drummond, 2007RAMBAUT, A. and DRUMMOND, A.J., 2007 [viewed 12 November 2017]. Tracer v. 1.4 [software]. Available from: http://beast.bio.ed.ac.uk/Tracer
http://beast.bio.ed.ac.uk/Tracer...
), in order to evaluate inter-specimen relationships (*BEAST) (Heled and Drummond, 2010HELED, J. and DRUMMOND, A.J., 2010. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution, vol. 27, no. 3, pp. 570-580. http://dx.doi.org/10.1093/molbev/msp274. PMid:19906793.
http://dx.doi.org/10.1093/molbev/msp274...
). Runs comprising 80 million generations were conducted, with samples being retrieved every 10,000 generations, of which 10% were discarded as burn-in. The runs were analyzed for possible cuts, using Tracer v.1.6, available in the BEAST program (Rambaut and Drummond, 2007RAMBAUT, A. and DRUMMOND, A.J., 2007 [viewed 12 November 2017]. Tracer v. 1.4 [software]. Available from: http://beast.bio.ed.ac.uk/Tracer
http://beast.bio.ed.ac.uk/Tracer...
), to verify whether the Effective Sample Size (ESS) was adequate, in all cases. All the trees generated in the present study were edited in Figtree v.1.3.1.
3. Results
The database included 477 base pairs (bps) for the 16S gene, 615 bps for COI, and 295 bps for Rhodopsin. The concatenated 16S+Rhod database contained 772 bps.
The greatest genetic distance was recorded for the COI marker (Table 2), with a divergence of 12.7% between S. nebulosus and S. constrictus. In the case of the 16S gene (Table 3), the divergence was of 4.7%, whereas for the Rhodopsin gene, the genetic distance between the two species was of only 1% (Table 4).
The same evolutionary model was selected by jModelTest 2 for the COI and Rhodopsin genes, with a different model selected for the 16S gene (Table 5). As the topologies retrieved by both analytical approaches (BI and ML) were highly consistent and very similar to one another, only the BI trees are presented herein. The ML trees can be provided upon request.
Evolutionary models used for the phylogenetic reconstructions applying the BI and ML methods.
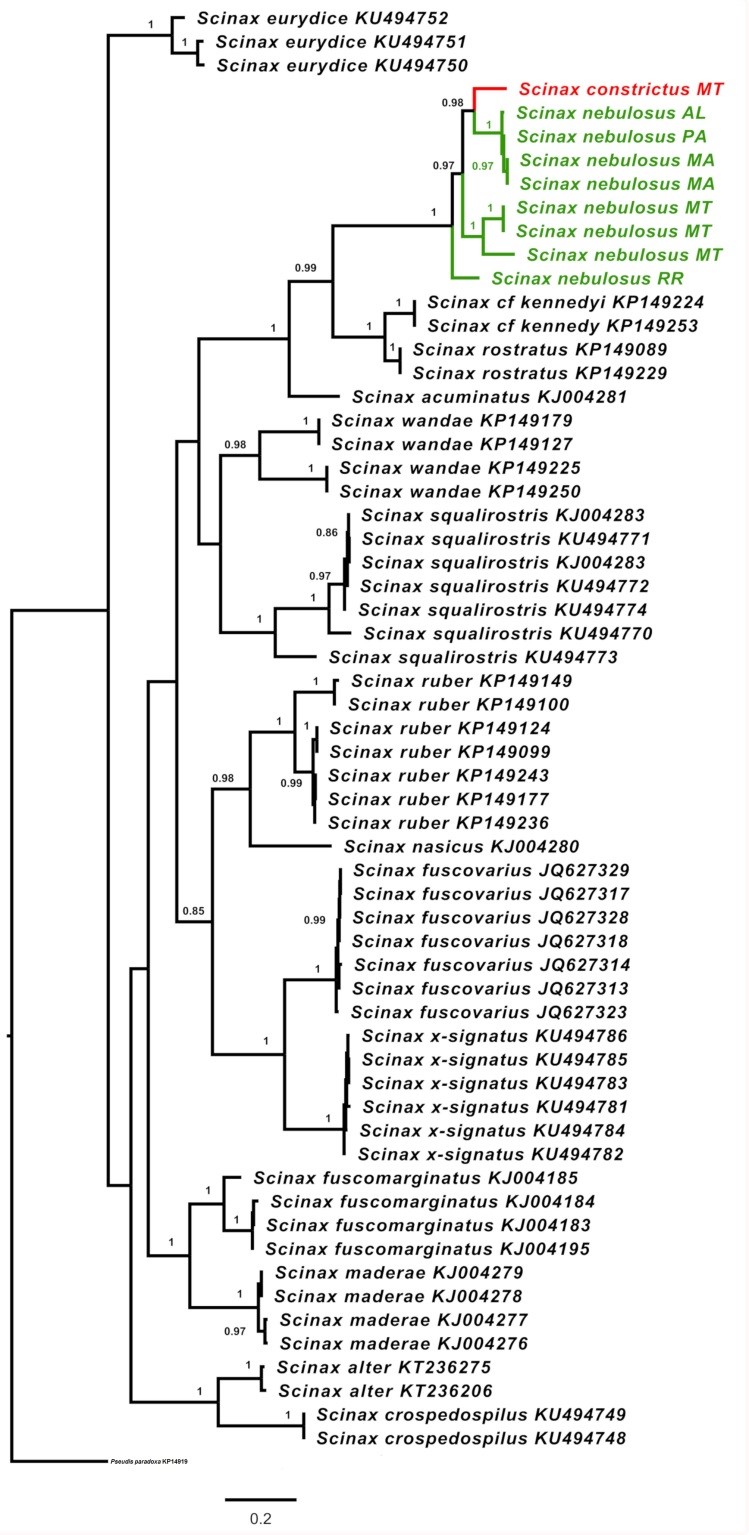
Scinax was monophyletic in all topologies derived from all different genetic markers, and in all analytical approaches. Regarding the BI phylogenetic tree obtained for the 16S marker, a number of the Scinax species also formed well-supported monophyletic groups. In this topology, S. nebulosus and S. constrictus form a well-supported clade in which both species tend to form natural groups (Figure 2). Poorly-identified specimens (labeled as S. gr. nebulosus) were grouped with either S. nebulosus or S. constrictus. Some structuring was observed for S. nebulosus samples. Specimens from northeastern (Alagoas and Maranhão) and central (Goiás) Brazil were separated with high support from both the BI (0.97) and ML (98%) analyses, as were those from two central Brazilian Cerrado portions, namely Mato Grosso and Tocantins (BI = 1.00, ML = 99%) and Mato Grosso alone (BI = 1.00, ML = 99%).
Tree obtained from the 16S gene analysis with significant bootstrap in the BI and ML analyses, respectively. Species name + GenBank voucher. The numbers on the branches represent the Bayesian (BI) support and bootstrap values (ML), respectively. Each color represents a monophyletic clade. The outgroups and paraphyletic groups are shown in black, with S. nebulosus in green and S. constrictus in red.
Scinax constrictus appears as a sister species for S. nebulosus in both the BI and ML analyses. Similar findings were obtained in the COI trees (Figure 3), in which S. constrictus and S. nebulosus appear as genetically distinct species, with high support values for the phylogenetic analyses (BI = 0.98, ML = 100%). As in the 16S trees, the COI tree recovered S. nebulosus as a polyphyletic species, formed by at least three groups, with S. constrictus as a sister group. One group contained the specimen from Roraima, the second group encompassed the specimens from Mato Grosso, while the third group included the sequences from Alagoas, Pará, and Maranhão.
Tree obtained from the COI gene analysis with significant bootstrap in the BI and ML analyses, respectively. Species name + GenBank voucher. The numbers on the branches represent the Bayesian (BI) support and bootstrap values (ML), respectively. Each color represents a monophyletic clade. The outgroups and paraphyletic groups are shown in black, S. nebulosus in green and S. constrictus in red.
The species tree obtained from BEAST also indicates that S. constrictus and S. nebulosus are separate genetic lineages, with high support values (1). The analysis nevertheless confirmed the close phylogenetic relationship between both species (Figure 4).
Species tree for the Scinax genus based on the concatenated data for the 16S and Rhodopsin genes. Values located at the nodes represent posterior probabilities, with only values equal to or above 0.99 shown.
4. Discussion
The genetic distances obtained from all three markers indicate that S. constrictus and S. nebulosus are distinct species. The divergence value recorded for the COI gene was of over 10%, a comparatively high value for differences between amphibian species (Vences et al., 2005aVENCES, M., THOMAS, M., BONETT, R.M. and VIEITES, D.R., 2005a. Deciphering amphibian diversity through DNA barcoding: chances and challenges. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, vol. 360, no. 1462, pp. 1859-1868. http://dx.doi.org/10.1098/rstb.2005.1717. PMid:16221604.
http://dx.doi.org/10.1098/rstb.2005.1717...
; Guarnizo et al., 2015GUARNIZO, C.E., PAZ, A., MUNOZ-ORTIZ, A., FLECHAS, S.V., MENDEZ-NARVAEZ, J. and CRAWFORD, A.J., 2015. DNA Barcoding Survey of Anurans across the Eastern Cordillera of Colombia and the Impact of the Andes on Cryptic Diversity. PLoS One, vol. 10, no. 5, pp. e0127312. http://dx.doi.org/10.1371/journal.pone.0127312. PMid:26000447.
http://dx.doi.org/10.1371/journal.pone.0...
). This differentiation was also supported by more conservative markers used in other studies (Nair et al., 2012NAIR, A., GOPALAN, S.V., GEORGE, S., KUMAR, K.S., TEACHER, A.G.F. and MERILA, J., 2012. High cryptic diversity of endemic Indirana frogs in the Western Ghats biodiversity hotspot. Animal Conservation, vol. 15, no. 5, pp. 489-498. http://dx.doi.org/10.1111/j.1469-1795.2012.00539.x.
http://dx.doi.org/10.1111/j.1469-1795.20...
; Nogueira et al., 2016NOGUEIRA, L., SOLÉ, M., SIQUEIRA, S., AFFONSO, P.R.A.M., STRÜSSMANN, C. and SAMPAIO, I., 2016. Genetic analysis reveals candidate species in the Scinax catharinae clade (Amphibia: Anura) from Central Brazil. Genetics and Molecular Biology, vol. 39, no. 1, pp. 49-53. http://dx.doi.org/10.1590/1678-4685-GMB-2015-0037. PMid:27007898.
http://dx.doi.org/10.1590/1678-4685-GMB-...
) and in the present study, i.e., 16S and Rhodopsin.
Based on an experimental analysis, Vences et al. (2005b)VENCES, M., THOMAS, M., VAN DER MEIJDEN, A., CHIARI, Y. and VIEITES, D.R., 2005b. Comparative performance of the 16S rRNA gene in DNA barcoding of amphibians. Frontiers in Zoology, vol. 2, no. 1, pp. 5. http://dx.doi.org/10.1186/1742-9994-2-5. PMid:15771783.
http://dx.doi.org/10.1186/1742-9994-2-5...
concluded that the mitochondrial 16S rRNA gene satisfies the prerequisites for a universal DNA barcode marker for amphibians and may be superior to COI sequences for the identification of vertebrate clades. This study observed that, while COI varied considerably in amphibians, 16S was highly conserved in these vertebrates. The authors thus recommend the use of 16S as an additional marker for vertebrate DNA barcoding.
Xia et al. (2012)XIA, Y., GU, H., PENG, R., CHEN, Q., ZHENG, Y., MURPHY, R.W. and ZENG, X., 2012. COI is better than 16S rRNA for DNA barcoding Asiatic salamanders (Amphibia: Caudata: Hynobiidae). Molecular Ecology Resources, vol. 12, no. 1, pp. 48-56. http://dx.doi.org/10.1111/j.1755-0998.2011.03055.x. PMid:21824335.
http://dx.doi.org/10.1111/j.1755-0998.20...
also support the use of the 16S rRNA gene for amphibian DNA barcoding, given that the authors indicated that genetic distances based on the Kimura 2-parameter (K2P) model revealed that the mean intraspecific distance was 1.4% in the COI gene, but only 0.3% in the 16S rRNA. The level of 16S rRNA variation obtained in the present study was higher than this aforementioned value, indicating a high degree of genetic variation among the study species, reinforcing their taxonomic validity.
Since the description of S. constrictus (Lima et al., 2004LIMA, L.P., BASTOS, R.P. and GIARETTA, A.A., 2004. A new Scinax Wagler, 1830 of the rostratus group from Central Brasil (Amphibia, Anura, Hylidae). Arquivos do Museu Nacional. Museu Nacional, vol. 62, pp. 505-512.), many specimens of both species have been difficult to classify reliably, given the lack of precision in the traditional morphological approach for taxa differentiation. In general, taxa with uncertain morphological variations are considered to be species complexes, with cryptic forms. This is probably the case for both S. nebulosus and S. constrictus, which present high levels of genetic differentiation in morphologically similar forms. In such cases, the molecular approach is essential for reliable species differentiation, as in the case of the Rhinella marina species complex, comprising distinct lineages composed of cryptic species (Bessa-Silva et al., 2016BESSA-SILVA, A.R., VALLINOTO, M., SODRÉ, D., CUNHA, D.B., ASP, N.E., SAMPAIO, I., SCHNEIDER, H. and SEQUEIRA, F., 2016. Patterns of genetic variability in Island Populations of the Cane Toad (Rhinella marina) from the Mouth of the Amazon. PLoS One, vol. 11, no. 4, pp. e0152492. http://dx.doi.org/10.1371/journal.pone.0152492. PMid:27073849.
http://dx.doi.org/10.1371/journal.pone.0...
).
In addition to confirming Scinax as a monophyletic group, the topologies obtained herein support the validation of S. constrictus and S. nebulosus as distinct species. Some aspects of the arrangement within some of the clades were unexpected, however, which could invalidate the species as a monophyletic group. Most of the incongruent samples were specimens recently assigned to the S. nebulosus group, probably as a consequence of the unreliable morphological definition of the species. Scinax constrictus can be distinguished from S. nebulosus by its smaller size, the presence of a discrete anal flap in males (absent in S. nebulosus), and the presence of a row of small tubercles on the lower jaw margin, which is absent or indistinct in S. nebulosus (Lima et al., 2004LIMA, L.P., BASTOS, R.P. and GIARETTA, A.A., 2004. A new Scinax Wagler, 1830 of the rostratus group from Central Brasil (Amphibia, Anura, Hylidae). Arquivos do Museu Nacional. Museu Nacional, vol. 62, pp. 505-512.). The advertisement call of S. constrictus is also quite distinct concerning dominant frequency and number of notes (Lima et al., 2004LIMA, L.P., BASTOS, R.P. and GIARETTA, A.A., 2004. A new Scinax Wagler, 1830 of the rostratus group from Central Brasil (Amphibia, Anura, Hylidae). Arquivos do Museu Nacional. Museu Nacional, vol. 62, pp. 505-512.), even though these traits vary considerably in S. nebulosus. Despite these diagnoses, the analysis of specimens diagnosed previously as S. nebulosus or S. constrictus indicate that these traits are not adequate for reliable species distinction (Weber, L.N. personal communication).
It is, therefore, clear, that morphological analyses are extremely vulnerable to error and taxa misidentification, made apparent by molecular tools. The geographic distributions of both taxa appear to support the molecular diagnosis, given that S. constrictus appears to be restricted to the Cerrado savanna of central Brazil, whereas S. nebulosus is more widely distributed, and may even be sympatric with S. constrictus.
Genetic analyses have been widely applied to validate anuran species described on the basis of morphological characteristics. Bruschi et al. (2013)BRUSCHI, D.P., BUSIN, C.S., TOLEDO, L.F., VASCONCELLOS, G.A., STRUSSMANN, C., WEBER, L.N., LIMA, A.P., LIMA, J.D. and RECCO-PIMENTEL, S.M., 2013. Evaluation of the taxonomic status of populations assigned to Phyllomedusa hypochondrialis (Anura, Hylidae, Phyllomedusinae) based on molecular, chromosomal, and morphological approach. BMC Genetics, vol. 14, no. 1, pp. 70. http://dx.doi.org/10.1186/1471-2156-14-70. PMid:23937545.
http://dx.doi.org/10.1186/1471-2156-14-7...
, for example, evaluated the taxonomic status of Phyllomedusa hypochondrialis (Anura, Hylidae, Phyllomedusinae) applying cytogenetic and molecular data to differentiate this species from Phyllomedusa azurea. Their results contributed to the extension of the geographic range of P. hypochondrialis, and confirmed its sympatry with P. azurea, given that incorrect specimen identification appeared to have resulted in inconsistencies in the original P. azurea geographic distribution definition. In the present study, the analysis of the 16S gene (Figure 2) indicated the grouping of S. nebulosus and S. constrictus specimens with S. constrictus specimens originally assigned to the nebulosus group, which appears to have been the result of species misidentification. In the case of both PhyllomedusaBruschi et al. (2013)BRUSCHI, D.P., BUSIN, C.S., TOLEDO, L.F., VASCONCELLOS, G.A., STRUSSMANN, C., WEBER, L.N., LIMA, A.P., LIMA, J.D. and RECCO-PIMENTEL, S.M., 2013. Evaluation of the taxonomic status of populations assigned to Phyllomedusa hypochondrialis (Anura, Hylidae, Phyllomedusinae) based on molecular, chromosomal, and morphological approach. BMC Genetics, vol. 14, no. 1, pp. 70. http://dx.doi.org/10.1186/1471-2156-14-70. PMid:23937545.
http://dx.doi.org/10.1186/1471-2156-14-7...
and Scinax (present study), molecular analyses indicated clearly a monophyletic position within the Hylidae family.
Brusquetti et al. (2014)BRUSQUETTI, F., JANSEN, M., BARRIO-AMORÓS, C., SEGALLA, M. and HADDAD, C.F.B., 2014. Taxonomic review of Scinax fuscomarginatus (Lutz, 1925) and related species (Anura; Hylidae). Zoological Journal of the Linnean Society, vol. 171, no. 4, pp. 783-821. http://dx.doi.org/10.1111/zoj.12148.
http://dx.doi.org/10.1111/zoj.12148...
reviewed the taxonomy of Scinax fuscomarginatus (Lutz, 1925), and evaluated morphologically similar species exhibiting controversial taxonomy, such as S. fuscumarginatus, S. parkeri, S. trilineatus, S. lutzorum and S. pusillus, by molecular analyses, with paraphyletic clades being recuperated for S. fuscomarginatus in comparison to the other species, similar to the paraphyletism between S. nebulosus and S. constrictus reported in the present study (Figures 2 and 3). Duellman et al. (2016)DUELLMAN, W.E., MARION, A.B. and HEDGES, S.B., 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa, vol. 4104, no. 1, pp. 1-109. http://dx.doi.org/10.11646/zootaxa.4104.1.1. PMid:27394762.
http://dx.doi.org/10.11646/zootaxa.4104....
subsequently redistributed Scinax species, and among the species analyzed by Brusquetti et al. (2014)BRUSQUETTI, F., JANSEN, M., BARRIO-AMORÓS, C., SEGALLA, M. and HADDAD, C.F.B., 2014. Taxonomic review of Scinax fuscomarginatus (Lutz, 1925) and related species (Anura; Hylidae). Zoological Journal of the Linnean Society, vol. 171, no. 4, pp. 783-821. http://dx.doi.org/10.1111/zoj.12148.
http://dx.doi.org/10.1111/zoj.12148...
, only S. fuscomarginatus was maintained in the genus.
In their phylogenetic and biogeographic analyses of the Hylidae family, Duellman et al. (2016)DUELLMAN, W.E., MARION, A.B. and HEDGES, S.B., 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa, vol. 4104, no. 1, pp. 1-109. http://dx.doi.org/10.11646/zootaxa.4104.1.1. PMid:27394762.
http://dx.doi.org/10.11646/zootaxa.4104....
proposed major changes in the Scinax genus arrangement, ranging from a new subfamily to species redistribution. Scinax nebulosus and S. constrictus remained in the genus, and the results of the present study confirm their status as valid Scinax genus lineages.
A number of the species described in the Scinax Wagler (Anura: Hylidae) genus require genetic validation, and the application of molecular markers has not only contributed to the identification of new species, but also in the differentiation of cryptic lineages (Nunes et al., 2012NUNES, I., KWET, A. and POMBAL, J.R., 2012. Taxonomic revision of the Scinax alter species complex (Anura: hylidae). Copeia, vol. 2012, no. 3, pp. 554-569. http://dx.doi.org/10.1643/CH-11-088.
http://dx.doi.org/10.1643/CH-11-088...
; Lourenço et al., 2013LOURENÇO, A.C.C., CARVALHO, A.L.G., BAÊTA, D., PEZZUTI, T.L. and LEITE, F.S.F., 2013. A new species of the Scinax catharinae group (Anura, Hylidae) from Serra da Canastra, southwestern state of Minas Gerais, Brazil. Zootaxa, vol. 3613, no. 6, pp. 573-588. http://dx.doi.org/10.11646/zootaxa.3613.6.4. PMid:24698839.
http://dx.doi.org/10.11646/zootaxa.3613....
; Sturaro and Peloso, 2014STURARO, M.J. and PELOSO, P.L.V., 2014. New species of Scinax Wagler, 1830 (Anura: Hylidae) From the Middle Amazon River Basin, Brazil. Papéis Avulsos de Zoologia, vol. 54, no. 2, pp. 9-23. http://dx.doi.org/10.1590/0031-1049.2014.54.02.
http://dx.doi.org/10.1590/0031-1049.2014...
; Nogueira et al., 2016NOGUEIRA, L., SOLÉ, M., SIQUEIRA, S., AFFONSO, P.R.A.M., STRÜSSMANN, C. and SAMPAIO, I., 2016. Genetic analysis reveals candidate species in the Scinax catharinae clade (Amphibia: Anura) from Central Brazil. Genetics and Molecular Biology, vol. 39, no. 1, pp. 49-53. http://dx.doi.org/10.1590/1678-4685-GMB-2015-0037. PMid:27007898.
http://dx.doi.org/10.1590/1678-4685-GMB-...
).
The molecular phylogeny of Fouquet et al. (2007)FOUQUET, A., VENCES, M., SALDUCCI, M.D., MEYER, A., MARTY, C., BLANC, M. and GILLES, A., 2007. Revealing cryptic diversity using molecular phylogenetics and phylogeography in frogs of the Scinax ruber and Rhinella margaritifera species groups. Molecular phylogenetics and evolution, vol. 43, no. 2, pp. 567-582. http://dx.doi.org/10.1016/j.ympev.2006.12.006. PMid:17303441.
http://dx.doi.org/10.1016/j.ympev.2006.1...
indicates that Scinax ruber, found in the French Guiana region, is represented by six divergent lineages. The molecular phylogeny obtained by Lourenço et al. (2013)LOURENÇO, A.C.C., CARVALHO, A.L.G., BAÊTA, D., PEZZUTI, T.L. and LEITE, F.S.F., 2013. A new species of the Scinax catharinae group (Anura, Hylidae) from Serra da Canastra, southwestern state of Minas Gerais, Brazil. Zootaxa, vol. 3613, no. 6, pp. 573-588. http://dx.doi.org/10.11646/zootaxa.3613.6.4. PMid:24698839.
http://dx.doi.org/10.11646/zootaxa.3613....
confirmed that the Scinax specimens collected at the Chapada dos Guimarães National Park, in central Brazil, which are morphologically similar to species from the S. catharinae group (sensuFaivovich et al., 2005FAIVOVICH, J., HADDAD, C.F.B., GARCIA, P.C.A., FROST, D.R., CAMPBELL, J.A. and WHEELER, W.C., 2005. Systematic review of the frog family Hylidae, with special reference to Hylinae: phylogenetic analysis and taxonomic revision. Bulletin of the American Museum of Natural History, vol. 294, no. 1, pp. 1-240. http://dx.doi.org/10.1206/0003-0090(2005)294[0001:SROTFF]2.0.CO;2.
http://dx.doi.org/10.1206/0003-0090(2005...
), do, in fact, represent a new species. Nogueira et al. (2016)NOGUEIRA, L., SOLÉ, M., SIQUEIRA, S., AFFONSO, P.R.A.M., STRÜSSMANN, C. and SAMPAIO, I., 2016. Genetic analysis reveals candidate species in the Scinax catharinae clade (Amphibia: Anura) from Central Brazil. Genetics and Molecular Biology, vol. 39, no. 1, pp. 49-53. http://dx.doi.org/10.1590/1678-4685-GMB-2015-0037. PMid:27007898.
http://dx.doi.org/10.1590/1678-4685-GMB-...
emphasize the need for further comparative analyses between these species, integrating morphological and bioacoustics data.
In this context, many researchers have highlighted the importance of a multiple approach, i.e., integrating molecular, morphological, bioacoustic, and ecological data, in taxonomic anuran assessments, in facilitating species identification (Padial et al., 2014PADIAL, J., GRANT, T. and FROST, D.R., 2014. Molecular systematics of terraranas (Anura: Brachycephaloidea) with an assessment of the effects of alignment and optimality criteria. Zootaxa, vol. 3825, no. 1, pp. 1-132. http://dx.doi.org/10.11646/zootaxa.3825.1.1. PMid:24989881.
http://dx.doi.org/10.11646/zootaxa.3825....
; Silva, 2017SILVA, P.H., 2017. Investigação taxonômica em populações de Scinax perereca Pombal Jr, Haddad & Kasahara, 1995 (Anura: Hylidae). São José do Rio Preto: Universidade Estadual Paulista “Júlio de Mesquita Filho”, 69 p. Dissertação de Mestrado em Biologia Animal.; Pinto et al., 2019PINTO, B.J., COLLI, G.R., HIGHAM, T.E., RUSSELL, A.P., SCANTLEBURY, D.P., VITT, L.J. and GAMBLE, T., 2019. Population genetic structure and species delimitation of a widespread, Neotropical dwarf gecko. Molecular Phylogenetics and Evolution, vol. 133, pp. 54-66. http://dx.doi.org/10.1016/j.ympev.2018.12.029. PMid:30590108.
http://dx.doi.org/10.1016/j.ympev.2018.1...
). Overall, molecular analyses aided in differentiating populations diagnosed as S. constrictus and S. nebulosus, despite the morphological similarities of both species.
Acknowledgements
We are especially grateful to all the scientific collections that provided samples for analysis in the present study, and would also like to thank the students that collected specimens in the field. We are also grateful to FAPEMA for financial support through the Universal Research Program. This study is part of TMBF’s doctoral thesis.
-
(With 4 figures)
References
- BESSA-SILVA, A.R., VALLINOTO, M., SODRÉ, D., CUNHA, D.B., ASP, N.E., SAMPAIO, I., SCHNEIDER, H. and SEQUEIRA, F., 2016. Patterns of genetic variability in Island Populations of the Cane Toad (Rhinella marina) from the Mouth of the Amazon. PLoS One, vol. 11, no. 4, pp. e0152492. http://dx.doi.org/10.1371/journal.pone.0152492 PMid:27073849.
» http://dx.doi.org/10.1371/journal.pone.0152492 - BOSSUYT, F. and MILINKOVITCH, M.C., 2000. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proceedings of the National Academy of Sciences of the United States of America, vol. 97, no. 12, pp. 6585-6590. http://dx.doi.org/10.1073/pnas.97.12.6585 PMid:10841558.
» http://dx.doi.org/10.1073/pnas.97.12.6585 - BRUSCHI, D.P., BUSIN, C.S., TOLEDO, L.F., VASCONCELLOS, G.A., STRUSSMANN, C., WEBER, L.N., LIMA, A.P., LIMA, J.D. and RECCO-PIMENTEL, S.M., 2013. Evaluation of the taxonomic status of populations assigned to Phyllomedusa hypochondrialis (Anura, Hylidae, Phyllomedusinae) based on molecular, chromosomal, and morphological approach. BMC Genetics, vol. 14, no. 1, pp. 70. http://dx.doi.org/10.1186/1471-2156-14-70 PMid:23937545.
» http://dx.doi.org/10.1186/1471-2156-14-70 - BRUSQUETTI, F., JANSEN, M., BARRIO-AMORÓS, C., SEGALLA, M. and HADDAD, C.F.B., 2014. Taxonomic review of Scinax fuscomarginatus (Lutz, 1925) and related species (Anura; Hylidae). Zoological Journal of the Linnean Society, vol. 171, no. 4, pp. 783-821. http://dx.doi.org/10.1111/zoj.12148
» http://dx.doi.org/10.1111/zoj.12148 - DARRIBA, D., TABOADA, G.L., DOALLO, R. and POSADA, D., 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, vol. 9, no. 8, pp. 772. http://dx.doi.org/10.1038/nmeth.2109 PMid:22847109.
» http://dx.doi.org/10.1038/nmeth.2109 - DUELLMAN, W.E., MARION, A.B. and HEDGES, S.B., 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa, vol. 4104, no. 1, pp. 1-109. http://dx.doi.org/10.11646/zootaxa.4104.1.1 PMid:27394762.
» http://dx.doi.org/10.11646/zootaxa.4104.1.1 - FAIVOVICH, J., HADDAD, C.F.B., GARCIA, P.C.A., FROST, D.R., CAMPBELL, J.A. and WHEELER, W.C., 2005. Systematic review of the frog family Hylidae, with special reference to Hylinae: phylogenetic analysis and taxonomic revision. Bulletin of the American Museum of Natural History, vol. 294, no. 1, pp. 1-240. http://dx.doi.org/10.1206/0003-0090(2005)294[0001:SROTFF]2.0.CO;2
» http://dx.doi.org/10.1206/0003-0090(2005)294[0001:SROTFF]2.0.CO;2 - FELSENSTEIN, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution; International Journal of Organic Evolution, vol. 39, no. 4, pp. 783-791. http://dx.doi.org/10.1111/j.1558-5646.1985.tb00420.x PMid:28561359.
» http://dx.doi.org/10.1111/j.1558-5646.1985.tb00420.x - FERRÃO, M., COLATRELI, O., FRAGA, R., KAEFER, I., MORAVEC, J. and LIMA, A.L., 2016. High Species Richness of Scinax Treefrogs (Hylidae) in a Threatened Amazonian Landscape Revealed by an Integrative Approach. PLoS One, vol. 11, no. 11, pp. e0165679. http://dx.doi.org/10.1371/journal.pone.0165679 PMid:27806089.
» http://dx.doi.org/10.1371/journal.pone.0165679 - FOLMER, O., BLACK, M., HOEH, W., LUTZ, R. and VRIJENHOEK, R., 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, vol. 3, no. 5, pp. 294-299. PMid:7881515.
- FOUQUET, A., VENCES, M., SALDUCCI, M.D., MEYER, A., MARTY, C., BLANC, M. and GILLES, A., 2007. Revealing cryptic diversity using molecular phylogenetics and phylogeography in frogs of the Scinax ruber and Rhinella margaritifera species groups. Molecular phylogenetics and evolution, vol. 43, no. 2, pp. 567-582. http://dx.doi.org/10.1016/j.ympev.2006.12.006 PMid:17303441.
» http://dx.doi.org/10.1016/j.ympev.2006.12.006 - FROST, D.R., 2017 [viewed 12 November 2017]. Amphibians species of the world 3.0: an online reference [online]. New York: American Museu of Natural History. Available from: http://research.amnh.org/herpetology/amphibia/index.php
» http://research.amnh.org/herpetology/amphibia/index.php - GUARNIZO, C.E., PAZ, A., MUNOZ-ORTIZ, A., FLECHAS, S.V., MENDEZ-NARVAEZ, J. and CRAWFORD, A.J., 2015. DNA Barcoding Survey of Anurans across the Eastern Cordillera of Colombia and the Impact of the Andes on Cryptic Diversity. PLoS One, vol. 10, no. 5, pp. e0127312. http://dx.doi.org/10.1371/journal.pone.0127312 PMid:26000447.
» http://dx.doi.org/10.1371/journal.pone.0127312 - GUINDON, S.J.F., DUFAYARD, V., LEFORT, M., ANISIMOVA, W., HORDIJK, W. and GASCUEL, O., 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, vol. 59, no. 3, pp. 307-321. http://dx.doi.org/10.1093/sysbio/syq010 PMid:20525638.
» http://dx.doi.org/10.1093/sysbio/syq010 - HELED, J. and DRUMMOND, A.J., 2010. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution, vol. 27, no. 3, pp. 570-580. http://dx.doi.org/10.1093/molbev/msp274 PMid:19906793.
» http://dx.doi.org/10.1093/molbev/msp274 - KATOH, K. and STANDLEY, D.M., 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Molecular Biology and Evolution, vol. 30, no. 4, pp. 772-780. http://dx.doi.org/10.1093/molbev/mst010 PMid:23329690.
» http://dx.doi.org/10.1093/molbev/mst010 - KUMAR, S., STECHER, G. and TAMURA, K., 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution, vol. 33, no. 7, pp. 1870-1874. http://dx.doi.org/10.1093/molbev/msw054 PMid:27004904.
» http://dx.doi.org/10.1093/molbev/msw054 - LIMA, L.P., BASTOS, R.P. and GIARETTA, A.A., 2004. A new Scinax Wagler, 1830 of the rostratus group from Central Brasil (Amphibia, Anura, Hylidae). Arquivos do Museu Nacional. Museu Nacional, vol. 62, pp. 505-512.
- LIMA, M.G., 2011. Variação geográfica de Scinax argyreornatus (Miranda- Ribeiro, 1926) (Anura: Hylidae) Recife: Universidade Federal de Pernambuco, 98 p. Dissertação de Mestrado em Biologia Animal.
- LOURENÇO, A.C.C., CARVALHO, A.L.G., BAÊTA, D., PEZZUTI, T.L. and LEITE, F.S.F., 2013. A new species of the Scinax catharinae group (Anura, Hylidae) from Serra da Canastra, southwestern state of Minas Gerais, Brazil. Zootaxa, vol. 3613, no. 6, pp. 573-588. http://dx.doi.org/10.11646/zootaxa.3613.6.4 PMid:24698839.
» http://dx.doi.org/10.11646/zootaxa.3613.6.4 - LOURENÇO, L.B., TARGUETA, C.P., BALDO, D., NASCIMENTO, J., GARCIA, P.C., ANDRADE, G.V., HADDAD, C.F. and RECCO-PIMENTEL, S.M., 2015. Phylogeny of frogs the genus Physalaemus (Anura, Leptodactylidae) inferred from mitochondrial and nuclear gene sequences. Molecular Phylogenetics and Evolution, vol. 92, pp. 204-216. http://dx.doi.org/10.1016/j.ympev.2015.06.011 PMid:26143292.
» http://dx.doi.org/10.1016/j.ympev.2015.06.011 - MENEZES, L., CANEDO, C., BATALHA-FILHO, H., GARDA, A.A., GEHARA, M. and NAPOLI, M.F., 2016. Multilocus phylogeography of the treefrog Scinax eurydice (Anura, Hylidae) reveals a plio-pleistocene diversification in the Atlantic Forest. PLoS One, vol. 11, no. 6, pp. e0154626. http://dx.doi.org/10.1371/journal.pone.0154626 PMid:27248688.
» http://dx.doi.org/10.1371/journal.pone.0154626 - NAIR, A., GOPALAN, S.V., GEORGE, S., KUMAR, K.S., TEACHER, A.G.F. and MERILA, J., 2012. High cryptic diversity of endemic Indirana frogs in the Western Ghats biodiversity hotspot. Animal Conservation, vol. 15, no. 5, pp. 489-498. http://dx.doi.org/10.1111/j.1469-1795.2012.00539.x
» http://dx.doi.org/10.1111/j.1469-1795.2012.00539.x - NICHOLS, R., 2001. Gene trees and species trees are not the same. Trends in Ecology & Evolution, vol. 16, no. 7, pp. 358-364. http://dx.doi.org/10.1016/S0169-5347(01)02203-0 PMid:11403868.
» http://dx.doi.org/10.1016/S0169-5347(01)02203-0 - NOGUEIRA, L., SOLÉ, M., SIQUEIRA, S., AFFONSO, P.R.A.M., STRÜSSMANN, C. and SAMPAIO, I., 2016. Genetic analysis reveals candidate species in the Scinax catharinae clade (Amphibia: Anura) from Central Brazil. Genetics and Molecular Biology, vol. 39, no. 1, pp. 49-53. http://dx.doi.org/10.1590/1678-4685-GMB-2015-0037 PMid:27007898.
» http://dx.doi.org/10.1590/1678-4685-GMB-2015-0037 - NUNES, I., KWET, A. and POMBAL, J.R., 2012. Taxonomic revision of the Scinax alter species complex (Anura: hylidae). Copeia, vol. 2012, no. 3, pp. 554-569. http://dx.doi.org/10.1643/CH-11-088
» http://dx.doi.org/10.1643/CH-11-088 - PADIAL, J., GRANT, T. and FROST, D.R., 2014. Molecular systematics of terraranas (Anura: Brachycephaloidea) with an assessment of the effects of alignment and optimality criteria. Zootaxa, vol. 3825, no. 1, pp. 1-132. http://dx.doi.org/10.11646/zootaxa.3825.1.1 PMid:24989881.
» http://dx.doi.org/10.11646/zootaxa.3825.1.1 - PALUMBI, S., MARTIN, A., ROMANO, S., MACMILLAN, W.O., STICE, L. and GRABOWSKI, G., 1991. The simple fool’s guide to PCR, Version 2.0 Honolulu: Department of Zoology and Kewalo Marine Laboratoly, University of Hawaii.
- PINTO, B.J., COLLI, G.R., HIGHAM, T.E., RUSSELL, A.P., SCANTLEBURY, D.P., VITT, L.J. and GAMBLE, T., 2019. Population genetic structure and species delimitation of a widespread, Neotropical dwarf gecko. Molecular Phylogenetics and Evolution, vol. 133, pp. 54-66. http://dx.doi.org/10.1016/j.ympev.2018.12.029 PMid:30590108.
» http://dx.doi.org/10.1016/j.ympev.2018.12.029 - PYRON, R.A. and WIENS, J.J., 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Molecular Phylogenetics and Evolution, vol. 61, no. 2, pp. 543-583. http://dx.doi.org/10.1016/j.ympev.2011.06.012 PMid:21723399.
» http://dx.doi.org/10.1016/j.ympev.2011.06.012 - RAMBAUT, A. and DRUMMOND, A.J., 2007 [viewed 12 November 2017]. Tracer v. 1.4 [software]. Available from: http://beast.bio.ed.ac.uk/Tracer
» http://beast.bio.ed.ac.uk/Tracer - RAMBAUT, A. and DRUMMOND, A.J., 2009 [viewed 12 November 2017]. Tracer version 1.5 [software]. Available from: http://beast.bio.ed.ac.uk
» http://beast.bio.ed.ac.uk - RONQUIST, F., TESLENKO, M., VAN DER MARK, P., AYRES, D.L., DARLING, A., HÖHNA, S., LARGET, B., LIU, L., SUCHARD, M.A. and HUELSENBECK, J.P., 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, vol. 61, no. 3, pp. 539-542. http://dx.doi.org/10.1093/sysbio/sys029 PMid:22357727.
» http://dx.doi.org/10.1093/sysbio/sys029 - SÁ, R.O., GRANT, T., CAMARGO, A., HEYER, R.W., PONSSA, M.L. and STANLEY, E., 2014. Systematics of the Neotropical Genus Leptodactylus Fitzinger, 1826 (Anura: Leptodactylidae): phylogeny, the relevance of non-molecular evidence, and species accounts São Paulo: Sociedade Brasileira de Herpetologia.
- SALDUCCI, M., MARTY, C., CHAPPAZ, R. and GILLES, A., 2002. Molecular phylogeny of French Guiana Hylinae: implications for the systematic and biodiversity of the Neotropical frogs. Comptes Rendus Biologies, vol. 325, no. 2, pp. 141-153. http://dx.doi.org/10.1016/S1631-0691(02)01423-3 PMid:11980175.
» http://dx.doi.org/10.1016/S1631-0691(02)01423-3 - SAMBROOK, J. and RUSSEL, D.W., 2001. Rapid isolation of yeast DNA. In: J. SAMBROOK and D.W. RUSSEL, eds. Molecular cloning, a laboratory manual New York: Cold Spring Harbor Laboratory, pp. 631-632.
- SILVA, P.H., 2017. Investigação taxonômica em populações de Scinax perereca Pombal Jr, Haddad & Kasahara, 1995 (Anura: Hylidae) São José do Rio Preto: Universidade Estadual Paulista “Júlio de Mesquita Filho”, 69 p. Dissertação de Mestrado em Biologia Animal.
- STURARO, M.J. and PELOSO, P.L.V., 2014. New species of Scinax Wagler, 1830 (Anura: Hylidae) From the Middle Amazon River Basin, Brazil. Papéis Avulsos de Zoologia, vol. 54, no. 2, pp. 9-23. http://dx.doi.org/10.1590/0031-1049.2014.54.02
» http://dx.doi.org/10.1590/0031-1049.2014.54.02 - SUK, H.Y., BAE, H.G., KIM, D.Y., WON, H., BAEK, H.J., LEE, C.H., KIM, D.Y., GO, Y.M., SONG, J.Y., LEE, H. and MIN, M.S., 2020. Genetic and phylogenetic structure of Hynobius quelpaertensis, an endangered endemic salamander species on the Korean Peninsula. Genes & Genomics, vol. 42, no. 2, pp. 165-178. PMid:31797315.
- TAKAZONE, A.M.G., 2015. Estudo taxonômico de Scinax nebulosus (SPIX, 1824) e Scinax constrictus (LIMA, BASTOS E GIARETTA, 2004) (ANURA, HYLIDAE) com base em parâmetros morfológicos, acústicos e de distribuição geográfica São Luís: Universidade Federal do Maranhão, 77 p. Dissertação de Mestrado em Biodiversidade e Conservação.
- VENCES, M., THOMAS, M., BONETT, R.M. and VIEITES, D.R., 2005a. Deciphering amphibian diversity through DNA barcoding: chances and challenges. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, vol. 360, no. 1462, pp. 1859-1868. http://dx.doi.org/10.1098/rstb.2005.1717 PMid:16221604.
» http://dx.doi.org/10.1098/rstb.2005.1717 - VENCES, M., THOMAS, M., VAN DER MEIJDEN, A., CHIARI, Y. and VIEITES, D.R., 2005b. Comparative performance of the 16S rRNA gene in DNA barcoding of amphibians. Frontiers in Zoology, vol. 2, no. 1, pp. 5. http://dx.doi.org/10.1186/1742-9994-2-5 PMid:15771783.
» http://dx.doi.org/10.1186/1742-9994-2-5 - XIA, Y., GU, H., PENG, R., CHEN, Q., ZHENG, Y., MURPHY, R.W. and ZENG, X., 2012. COI is better than 16S rRNA for DNA barcoding Asiatic salamanders (Amphibia: Caudata: Hynobiidae). Molecular Ecology Resources, vol. 12, no. 1, pp. 48-56. http://dx.doi.org/10.1111/j.1755-0998.2011.03055.x PMid:21824335.
» http://dx.doi.org/10.1111/j.1755-0998.2011.03055.x
Publication Dates
-
Publication in this collection
23 Oct 2020 -
Date of issue
Oct-Dec 2021
History
-
Received
26 June 2019 -
Accepted
04 May 2020