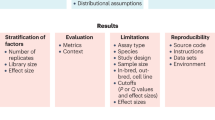
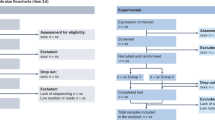
Abstract
Computational methods are key in microbiome research, and obtaining a quantitative and unbiased performance estimate is important for method developers and applied researchers. For meaningful comparisons between methods, to identify best practices and common use cases, and to reduce overhead in benchmarking, it is necessary to have standardized datasets, procedures and metrics for evaluation. In this tutorial, we describe emerging standards in computational meta-omics benchmarking derived and agreed upon by a larger community of researchers. Specifically, we outline recent efforts by the Critical Assessment of Metagenome Interpretation (CAMI) initiative, which supplies method developers and applied researchers with exhaustive quantitative data about software performance in realistic scenarios and organizes community-driven benchmarking challenges. We explain the most relevant evaluation metrics for assessing metagenome assembly, binning and profiling results, and provide step-by-step instructions on how to generate them. The instructions use simulated mouse gut metagenome data released in preparation for the second round of CAMI challenges and showcase the use of a repository of tool results for CAMI datasets. This tutorial will serve as a reference for the community and facilitate informative and reproducible benchmarking in microbiome research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The results of all benchmarked methods and gold standards are available at https://zenodo.org/communities/cami. Links to individual results and DOIs are available in Supplementary Tables 1, 4, 8, and 11. The gold-standard assembly is provided with the CAMI II mouse gut dataset (Table 2). Assembly results and code used to generate Fig. 3 are available at https://github.com/CAMI-challenge/BenchmarkingToolkitTutorial. Genome and taxonomic binning, and taxonomic profiling results used in Figs. 4–7 are available, respectively, in the AMBER and OPAL GitHub repositories at https://github.com/CAMI-challenge/AMBER and https://github.com/CAMI-challenge/OPAL. The code in this paper has been peer-reviewed.
References
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004).
Mitchell, A. L. et al. EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res 46, D726–D735 (2018).
Chen, I.-M. A. et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 47, D666–D677 (2019).
Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J. & Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35, 833–844 (2017).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019).
Almeida, A. et al. A new genomic blueprint of the human gut microbiota. Nature 568, 499–504 (2019).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Sczyrba, A. et al. Critical Assessment of Metagenome Interpretation—a benchmark of metagenomics software. Nat. Methods 14, 1063–1071 (2017).
Bansal, V. & Boucher, C. Sequencing technologies and analyses: where have we been and where are we going? iScience 18, 37–41 (2019).
Mantere, T., Kersten, S. & Hoischen, A. Long-read sequencing emerging in medical genetics. Front. Genet. 10, 426 (2019).
Mosimann, S., Meleshko, R. & James, M. N. A critical assessment of comparative molecular modeling of tertiary structures of proteins. Proteins 23, 301–317 (1995).
Andreoletti, G., Pal, L. R., Moult, J. & Brenner, S. E. Reports from the fifth edition of CAGI: The Critical Assessment of Genome Interpretation. Hum. Mutat. 40, 1197–1201 (2019).
Dessimoz, C., Škunca, N. & Thomas, P. D. CAFA and the open world of protein function predictions. Trends Genet 29, 609–610 (2013).
Weber, L. M. et al. Essential guidelines for computational method benchmarking. Genome Biol. 20, 125 (2019).
Mangul, S. et al. Systematic benchmarking of omics computational tools. Nat. Commun. 10, 1393 (2019).
Mavromatis, K. et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods 4, 495–500 (2007).
Lindgreen, S., Adair, K. L. & Gardner, P. P. An evaluation of the accuracy and speed of metagenome analysis tools. Sci. Rep. 6, 19233 (2016).
McIntyre, A. B. R. et al. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol. 18, 182 (2017).
Ye, S. H., Siddle, K. J., Park, D. J. & Sabeti, P. C. Benchmarking metagenomics tools for taxonomic classification. Cell 178, 779–794 (2019).
Bremges, A. & McHardy, A. C. Critical Assessment of Metagenome Interpretation enters the second round. mSystems 3, e00103-18 (2018).
Fritz, A. et al. CAMISIM: simulating metagenomes and microbial communities. Microbiome 7, 17 (2019).
Singer, E. et al. Next generation sequencing data of a defined microbial mock community. Sci. Data 3, 160081 (2016).
Mikheenko, A., Saveliev, V. & Gurevich, A. MetaQUAST: evaluation of metagenome assemblies. Bioinformatics 32, 1088–1090 (2016).
Meyer, F. et al. AMBER: Assessment of Metagenome BinnERs. GigaScience 7, giy069 (2018).
Meyer, F. et al. Assessing taxonomic metagenome profilers with OPAL. Genome Biol. 20, 51 (2019).
Grüning, B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods 15, 475–476 (2018).
Köster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Belmann, P. et al. Bioboxes: standardised containers for interchangeable bioinformatics software. Gigascience 4, 47 (2015).
da Veiga Leprevost, F. et al. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics 33, 2580–2582 (2017).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35, D61–D65 (2007).
McDonald, D. et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience 1, 7 (2012).
Li, D. et al. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11 (2016).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27, 824–834 (2017).
Mineeva, O., Rojas-Carulla, M., Ley, R. E., Schölkopf, B. & Youngblut, N. D. DeepMAsED: evaluating the quality of metagenomic assemblies. Bioinformatics 36, 3011–3017 (2020).
Clark, S. C., Egan, R., Frazier, P. I. & Wang, Z. ALE: a generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies. Bioinformatics 29, 435–443 (2013).
Kuhring, M., Dabrowski, P. W., Piro, V. C., Nitsche, A. & Renard, B. Y. SuRankCo: supervised ranking of contigs in de novo assemblies. BMC Bioinforma. 16, 240 (2015).
Wu, Y.-W., Simmons, B. A. & Singer, S. W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016).
Kang, D. D., Froula, J., Egan, R. & Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015).
Alneberg, J. et al. Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25, 1043–1055 (2015).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Wood, D. E. & Salzberg, S. L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014).
Gregor, I., Dröge, J., Schirmer, M., Quince, C. & McHardy, A. C. PhyloPythiaS+: a self-training method for the rapid reconstruction of low-ranking taxonomic bins from metagenomes. PeerJ 4, e1603 (2016).
von Meijenfeldt, F. A. B., Arkhipova, K., Cambuy, D. D., Coutinho, F. H. & Dutilh, B. E. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 20, 217 (2019).
Huson, D. H. et al. MEGAN Community Edition – interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12, e1004957 (2016).
Almeida, A. et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 39, 105–114 (2020).
Truong, D. T. et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015).
Milanese, A. et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 10, 1014 (2019).
Lu, J., Breitwieser, F. P., Thielen, P. & Salzberg, S. L. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3, e104 (2017).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018).
Ciccarelli, F. D. et al. Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287 (2006).
Konstantinidis, K. T. & Tiedje, J. M. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264 (2005).
McDonald, D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2012).
Segata, N. On the road to strain-resolved comparative metagenomics. mSystems 3, e00190-17 (2018).
Quince, C. et al. DESMAN: a new tool for de novo extraction of strains from metagenomes. Genome Biol. 18, 181 (2017).
Truong, D. T., Tett, A., Pasolli, E., Huttenhower, C. & Segata, N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27, 626–638 (2017).
Moss, E. L., Maghini, D. G. & Bhatt, A. S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat. Biotechnol. 38, 701–707 (2020).
Sajulga, R. et al. Survey of metaproteomics software tools for functional microbiome analysis. PLoS ONE 15, e0241503 (2020).
Acknowledgements
The authors thank P. B. Pope for helpful comments. A.E.D.’s contribution was facilitated in part by the Australian Research Council’s Discovery Projects funding scheme (project DP180101506). A.G.’s contribution was facilitated by St. Petersburg State University, Russia (grant ID PURE 51555639).
Author information
Authors and Affiliations
Contributions
F.M. and T.-R.L. performed the experiments; F.M., A.F., T.-R.L., and A.S. prepared the data; A.C.M., A.B., and A.S. conceived the experiments; A.C.M., F.M., and A.B. wrote the manuscript with comments by others; F.M., T.-R.L., D.K., A.F., A.G., A.E.D., A.S., A.B., and A.C.M. interpreted the results, and read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–13, Supplementary Figs. 1 and 2 and Supplementary Note.
Supplementary Data 1
Supplementary Results: MetaQUAST metrics
Rights and permissions
About this article
Cite this article
Meyer, F., Lesker, TR., Koslicki, D. et al. Tutorial: assessing metagenomics software with the CAMI benchmarking toolkit. Nat Protoc 16, 1785–1801 (2021). https://doi.org/10.1038/s41596-020-00480-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00480-3
This article is cited by
-
Challenges and best practices in omics benchmarking
Nature Reviews Genetics (2024)
-
Effective binning of metagenomic contigs using contrastive multi-view representation learning
Nature Communications (2024)
-
MetaBinner: a high-performance and stand-alone ensemble binning method to recover individual genomes from complex microbial communities
Genome Biology (2023)
-
Challenges and opportunities in sharing microbiome data and analyses
Nature Microbiology (2023)
-
How is Big Data reshaping preclinical aging research?
Lab Animal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.