Abstract
The outbreak of the new severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a public health emergency. Asthma does not represent a risk factor for COVID-19 in several published cohorts. We hypothesized that the SARS-CoV-2 proteome contains T cell epitopes, which are potentially cross-reactive to allergen epitopes. We aimed at identifying homologous peptide sequences by means of two distinct complementary bioinformatics approaches. Pipeline 1 included prediction of MHC Class I and Class II epitopes contained in the SARS-CoV-2 proteome and allergens along with alignment and elaborate ranking approaches. Pipeline 2 involved alignment of SARS-CoV-2 overlapping peptides with known allergen-derived T cell epitopes. Our results indicate a large number of MHC Class I epitope pairs including known as well as de novo predicted allergen T cell epitopes with high probability for cross-reactivity. Allergen sources, such as Aspergillus fumigatus, Phleum pratense and Dermatophagoides species are of particular interest due to their association with multiple cross-reactive candidate peptides, independently of the applied bioinformatic approach. In contrast, peptides derived from food allergens, as well as MHC class II epitopes did not achieve high in silico ranking and were therefore not further investigated. Our findings warrant further experimental confirmation along with examination of the functional importance of such cross-reactive responses.
Similar content being viewed by others
Introduction
The World Health Organization (WHO) has declared the outbreak of the new Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2, ssRNA virus, associated with COVID-19) as a public health emergency. As per the WHO report of 20 September 2020, more than 30 million cases and over 950,000 deaths have been reported worldwide1. Human coronaviruses are positive-sense single-stranded RNA (+ ssRNA) viruses, with SARS-CoV-2 and SARS-CoV belonging to the B-lineage of the Betacoronavirus genera and MERS-CoV to the C-lineage of the same genera2,3. The clinical features in patients affected with these respiratory viruses ranges from asymptomatic carriers to severe respiratory illness with pneumonia and acute respiratory distress syndrome (ARDS). In addition, a number of interesting vascular and inflammatory presentations have been noted, including a multisystem inflammatory syndrome in children.
We have previously reported on heterologous immune responses induced by influenza, another respiratory RNA virus, against allergens, which mediated protection from experimental allergic asthma4. Indeed, virus-induced T cell mediated heterologous immunity has been widely described in a variety of settings, which can confer protection or drive immunopathology against other antigens5,6. Given that the host immune response to SARS-CoV-2 and associated disease course can be so varied from patient to patient, this spectrum of presentations raises the question of what drives the differential host immune response. There is still little known about asthma phenotypes and severity of COVID-19. In general, asthma has not been shown to be a risk factor for COVID-19 in several published cohorts7,8. However, recent studies from the UK and the USA indicated higher numbers of asthmatics in COVID-19 patients9.
Interestingly, the UK Biobank recently reported that non-allergic patients had a higher risk of severe COVID-19, compared to patients with allergic asthma10. Moreover, evidence of T cell activation, as indicated by generation of effector (CD45RA-CD62L-) and central memory (CD45RA− CD62L+) αβCD4conv and CD8+ cells was reported among COVID-19 patients with mild or severe disease, suggesting that activation of T cells is inversely associated with severity of SARS-CoV-2 infection11. Other investigators reported that T cells in peripheral blood of COVID-19 patients with allergy were increased as compared to patients without allergies12. These preliminary clinical and laboratory observations along with our prior experimental evidence involving RNA viruses led us hypothesize that SARS-CoV-2 may share a degree of protein sequence homology to allergens, which may lead to the generation of cross-reactive T cell epitopes. Pre-existing T cells specific for such cross-reactive allergen-derived epitopes may have an impact on COVID-19 outcome via aberrant cytokine responses to the virus peptides. Indeed, these cytokines could prevent an overshooting T1 inflammatory reaction, both locally (as in the case of preexisting pulmonary CD4+ T cells specific to inhalant allergens) and/or systemically. Therefore, we sought to predict potentially cross-reactive allergen- and SARS-CoV-2-derived MHC Class I and Class II T cell epitopes, which can be presented by the most prevalent HLA alleles.
Methods and results
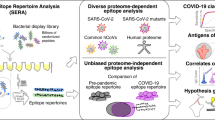
In order to examine our working hypothesis, we applied two distinct independent, complementary and systematic bioinformatics approaches (Fig. 1): (a) Pipeline 1-prediction of MHC Class I and Class II epitopes contained in the SARS-CoV-2 proteome and a comprehensive set of allergen protein sequences combined with alignment strategies and ranking of results based on clinical and sequence conservation criteria and (b) Pipeline 2- alignment of SARS-CoV-2 overlapping peptides with known allergen-derived T cell epitopes13.
Schematic overview of the bioinformatics approaches. A Pipeline 1; SARS-CoV-2 proteins were aligned against > 2500 allergen protein sequences (see methods) and MHC class I-and II- restricted potentially cross-reactive T cell epitope pairs were identified for the most frequent human HLA alleles. B Pipeline 2; In an independent framework, we performed the comparative analysis of sequential kmers from SARS-CoV-2 protein sequences with known IEDB allergen peptides to predict the cross-reactive viral peptide pool.
Pipeline 1
More than > 2500 allergen protein sequences were downloaded (dates of access 10.09.2017) from Allergome14,15,16,17 (Supplementary Table S1), and protein sequences for SARS-CoV-2 from UniProt18 (Supplementary Table S2). Viral T cell epitope prediction was performed using smm19, ann20 and consensus21 for MHC Class I (IC50 threshold < = 5000), and netMHCII22 for MHC Class II (affinity score threshold for strong binders: 0.500; for weak binders: 2.000) (Supplementary Table S3). Epitopes predicted by all methods were aligned against all allergen proteins using a local version of the NCBI protein blast program23. Allergen proteins associated with an alignment e-value < 10 were further processed for T cell epitope prediction using netMHC24 and netMHCpan25 for MHC Class I, and netMHCII and netMHCIIpan26 for MHC Class II prediction (affinity score threshold for strong binders: 0.500; for weak binders: 2.000). Viral and allergen epitopes were pairwise aligned with Biopython module pairwise 227 and for pairs with a score > 8, a final pair combined score (pcs) was calculated (Supplementary Methods). A higher pair combined score indicates a higher chance for a peptide pair to be cross-reactive and binding to MHC molecules. Duplicates among the resulting candidate epitope pairs were removed before further processing. Therefore, possible sequence repetition due to isoforms and isoallergens (Supplementary Table S1) do not influence further analyses. In total, we obtained more than 5000 candidate pairs for each, MHC Class I and Class II. The top 30 candidate epitope pairs, as per pair combined score, are listed for aero- and food allergens, MHC Class I and Class II presentation background in Supplementary Tables S4–S7, respectively. The top 30 MHC Class II restricted predicted virus-allergen pairs achieved relatively low pcs (24–657) as compared to Class I epitope pairs (1036–10,816). Among our top 30 MHC Class I potentially cross-reactive allergen derived epitopes, we identified more than 20 distinct protein families (Allfam database). In addition to MHC binding affinity and homology between peptide sequences, also other factors (e.g. conservation, association with clinical reactions) are important for the clinical relevance of peptides predicted to be cross-reactive at the T cell level. In order to capture this information level in our ranking, all allergen peptides and associated sources listed among the top 30 candidate epitope pairs were evaluated further with a scoring system (Supplementary Fig. S1 and Supplementary Methods). We found that the top 5 Class I aeroallergens were on average associated with higher pcs as compared to the top 5 potentially cross-reactive food allergens (Table 1 for MHC Class I and Table 2 for MHC Class II peptide pairs).
Pipeline 2
We obtained all known allergen-derived linear T cell epitope peptides from the IEDB, containing peptides known to bind MHC molecules with at least one published experimental evidence (e.g. based on the results of a T cell assay) (Supplementary Table S8). A total of 8207 antigenic peptides from 142 antigens were selected for evaluation, among which, peptides with ambiguous amino acids (e.g. with unknown amino acid ‘X’ or any special character) were removed from the subsequent analysis. Therefore, all included peptides could be defined in full. Next, SARS-CoV-2 protein sequences were analyzed for the potential antigenic regions by splitting each of the sequence into sequential k-mers (length = 15), and homology with allergen antigenic peptides was then profiled. Within a given threshold range, we found 43 unique SARS-CoV-2 peptides that belong to replicase poly protein and spike glycoprotein (Supplementary Table S9). These peptides demonstrate homology with antigenic peptides of 6 different allergen sources, including Canis lupus, Dermatophagoides farinae, Dermatophagoides pteronyssinus, Aspergillus fumigatus, Alternaria alternata and Phleum pratense, all of which are known to be respiratory allergens and, in the majority, clinically highly relevant (e.g. aeroallergens; Fig. 1).
However, despite the homology, it is likely that some of the peptides may not have strong MHC Class I binding affinity, and thus be less likely to be presented as antigens by HLA molecules. Therefore, we assessed the binding affinity of these peptides with human MHC Class I molecules, across a broad range of alleles that are known to bind viral proteins (52 most common HLA-A and HLA-B alleles). We observed that some of these peptides (n = 79) were predicted to have MHC Class I binding epitope regions associated with at least one of the Class I HLA alleles with IC50 < 500 nm (Supplementary Table S10). These antigenic peptides were predicted to bind with 20 most frequently occurring HLA Class I alleles, in which HLA*02:03 and HLA*02:06 were predicted to present the highest number of epitope residues. To further investigate if these peptides are specific to the coronavirus family, we performed the BLAST comparison with 2807 known viral antigenic peptides of bacteria, influenza-and corona- virus family (non-SARS CoV-2) from IEDB (with at least one T cell assay evidence) and filtered out matching peptides (Blast e-value < 1 and identity > 70%). Finally, we present 48 high-affinity HLA-binding peptides which are unique to the SARS-CoV-2 proteome, not common to bacteria, influenza and corona virus family antigenic peptides within a given threshold range (Supplementary Table S11) with 14 high confidence HLA Class I binding peptides with IC50 < 50 nm (Table 3).
Application of both complementary pipelines aimed at identifying T cell epitope pairs, which are highly likely to be cross-reactive. Pipeline 1 includes a broader approach by means of considering as many allergen protein sequences are available and subsequently predicting MHC binding affinity and performing alignment of the candidate epitopes. The elaborate scoring system which followed, prioritized these candidates based on clinical aspects and thus relevance. This pipeline has already been used and experimentally validated by our group for similar analyses (Balz K. et al., unpublished data). Nevertheless, immunogenicity and cross-reactivity of the peptides identified by pipeline 1 in our index work remains to be shown. Pipeline 2 takes only known immunogenic peptides into consideration, leading to more robust results. Further, alignment of these epitopes against the proteome of other organisms, allows identification of epitopes, which are unique for SARS-CoV-2. The approach of pipeline 2, however, does not allow identification of newly described T cell epitopes and less studied allergens are not considered.
Discussion
We have applied two independent, complementary and systematic bioinformatic approaches in order to identify potentially cross-reactive allergen- and SARS-CoV-2- T cell epitopes. Our in silico analysis revealed numerous candidate epitope pairs, including previously published and predicted peptides, while both applied pipelines highlighted an important role of MHC class I inhalant allergens. Epitope pairs including peptides from food allergens appeared to be of lower importance. Our finding may indicate that patients with respiratory allergies including asthma may be more affected by heterologous immune response against SARS-CoV-2. It is of high relevance, that both pipelines highlighted candidate epitopes from Dermatophagoides species, as well as Aspergillus fumigatus and Phleum pratense, suggesting an important role for these allergens. Although the frequency of allergen-specific CD8+ T cells is likely to be low, rare cell subsets have been quite often shown to play an important pathophysiological role28, and new technologies and bioinformatic approaches for identification of such populations are steadily emerging29. Quite importantly, the SARS-CoV-2 Nsp6141-149, which was identified among our top potentially cross-reactive epitope pairs, has been recently described by an independent group30. To our knowledge, this is the first report on in silico predicted T cell epitope cross-reactivity between SARS-CoV-2 and allergens. While a limitation of our study is the in silico nature of the work, the sequence homology between SARS-CoV-2 and clinically relevant respiratory allergens is along the lines of previously reported cross-reactivity between RNA virus- and allergen-derived peptides at the level of T memory cells4. Moreover, our current findings generate further hypotheses in how the adaptive immune system responds differentially with respect to the atopy status of the host. Our present study warrants an immediate investigation of these predicted T cell epitopes to link their possible role in driving the immune response against the SARS-CoV-2 and eventually shape COVID-19 outcome.
There are several different avenues through which the similarities may influence the host immune response. For instance, in hosts sensitized to one of the predicted aeroallergens, the identified similarities with the SARS-CoV-2 proteome may be protective if they prevent an overwhelming Th1 response and the accompanying cytokine storm. Furthermore, allergen-specific T cells may develop a memory response against heterologous SARS-CoV-2 epitopes, which is faster and more efficient. Conversely, such heterologous immune responses could have an adverse outcome by attenuating the antiviral response. T2 immune bias could potentially lead to inadequate virus clearance due to attenuated CD8+ responses. Indeed, there is evidence of a reciprocal relationship between atopy and production of type I and III Interferons in response to viral infections31. Given that underlying atopic conditions have not been identified as a significant risk factor for severe clinical courses in those infected with SARS-CoV-2, the epitope homology most likely plays a protective role7,8. Interestingly, Jackson et al32 recently reported that nasal epithelial cells from children with atopic asthma express significantly lower levels of ACE2 receptor as compared to cells from children without asthma or with non-atopic asthma. Similarly, another study using adult bronchial brush samples showed an inverse correlation between ACE2 gene expression and a Th2 dependent gene expression signature33. Differential expression of ACE2 receptors among atopic individuals could represent a distinct and unrelated mechanism of action in this context. Our in silico data provide ground to investigate the role of cellular immune responses in regards to the interaction between atopy/asthma and COVID-19. Indeed, the role of SARS-CoV-2-specific T cells in exposed and non-exposed individuals, thereby underlining the importance of heterologous immunity, has been very recently described34,35. Further experimental studies are needed to explore the involved pathogenetic mechanisms and potential clinical implications of underlying aeroallergen sensitization on the immune response to SARS-CoV-2.
Data availability
The data used and analyzed in the present study are available from the corresponding author on reasonable request.
Abbreviations
- ACE2:
-
Angiotensin converting enzyme 2
- ARDS:
-
Acute respiratory distress syndrome
- BLAST:
-
Basic local alignment search tool
- HLA:
-
Human leukocyte antigen
- IEDB:
-
Immune epitope database
- MHC:
-
Major histocompatibility complex
- NCBI:
-
National Center for Biotechnology Information
- RNA:
-
Ribonucleic acid
- +ssRNA:
-
Positive-sense single-stranded RNA
- Th 1:
-
T-helper 1
- Th 2:
-
T-helper 2
References
Coronavirus Disease (COVID-19) Situation Reports; 22.09.2020. [Cited 2020 September 22.] Available from https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/.
Nextstrain / groups / blab / sars-like-cov; 21.09.2020. [Cited 2020 September 22.] Available from https://nextstrain.org/groups/blab/sars-like-cov.
Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S. C. & Di Napoli, R. StatPearls. Features, Evaluation, and Treatment of Coronavirus (COVID-19). Treasure Island (FL) (2020).
Skevaki, C. et al. Influenza-derived peptides cross-react with allergens and provide asthma protection. J. Allergy Clin. Immunol. 142, 804–814 (2018).
Balz, K., Trassl, L., Härtel, V., Nelson, P. P. & Skevaki, C. Virus-induced T cell-mediated heterologous immunity and vaccine development. Front. Immunol. 11, 513 (2020).
Pusch, E., Renz, H. & Skevaki, C. Respiratory virus-induced heterologous immunity: Part of the problem or part of the solution?. Allergo J. Interdiszipl. Z. Allergol. Umweltmed. Organ Dtsch. Ges. Allerg. Immunitatsforschung 27, 28–45 (2018).
Liu, S., Zhi, Y. & Ying, S. COVID-19 and asthma: Reflection during the pandemic. Clin. Rev. Allergy Immunol. 59, 78–88 (2020).
Carli, G., Cecchi, L., Stebbing, J., Parronchi, P., Farsi, A. Is asthma protective against COVID-19? Allergy. https://doi.org/10.1111/all.14426 (2020).
Asthma Prevalence; 22.09.2020. [Cited 2020 September 22.] Available from https://www.cdc.gov/asthma/data-visualizations/prevalence.htm.
Zhu, Z. et al. Association of asthma and its genetic predisposition with the risk of severe COVID-19. J. Allergy Clin. Immunol. 146(327–329), e4 (2020).
Odak, I. et al. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine 57, 102885 (2020).
Shi, W. et al. Clinical characteristics of COVID-19 patients combined with allergy. Allergy 75, 2405–2408 (2020).
Vita, R. et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2019).
Mari, A., Rasi, C., Palazzo, P. & Scala, E. Allergen databases: current status and perspectives. Curr. Allergy Asthma Rep. 9, 376–383 (2009).
Radauer, C., Bublin, M., Wagner, S., Mari, A. & Breiteneder, H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 121(847–52), e7 (2008).
Cui, J. et al. Computer prediction of allergen proteins from sequence-derived protein structural and physicochemical properties. Mol. Immunol. 44, 514–520 (2007).
Mari, A. et al. Bioinformatics applied to allergy: allergen databases, from collecting sequence information to data integration. The Allergome platform as a model. Cell. Immunol. 244, 97–100 (2006).
The universal protein resource (UniProt). Nucleic Acids Res. 36: D190–D195 (2008).
Peters, B. & Sette, A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinform. 6, 132 (2005).
Buus, S. et al. Sensitive quantitative predictions of peptide-MHC binding by a “Query by Committee” artificial neural network approach. Tissue Antigens 62, 378–384 (2003).
Moutaftsi, M. et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 24, 817–819 (2006).
Nielsen, M. & Lund, O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinform. 10, 296 (2009).
Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinform. 10, 421 (2009).
Nielsen, M. et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. Publ. Protein Soc. 12, 1007–1017 (2003).
Hoof, I. et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61, 1–13 (2009).
Andreatta, M. et al. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 67, 641–650 (2015).
Cock, P. J. A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics (Oxford, England) 25, 1422–1423 (2009).
Murray, S. E., Toren, K. G. & Parker, D. C. Peripheral CD4(+) T-cell tolerance is induced in vivo by rare antigen-bearing B cells in follicular, marginal zone, and B-1 subsets. Eur. J. Immunol. 43, 1818–1827 (2013).
Arvaniti, E. & Claassen, M. Sensitive detection of rare disease-associated cell subsets via representation learning. Nat. Commun. 8, 14825 (2017).
Grifoni, A. et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 27(671–680), e2 (2020).
Edwards, M. R. et al. Viral infections in allergy and immunology: How allergic inflammation influences viral infections and illness. J. Allergy Clin. Immunol. 140, 909–920 (2017).
Jackson, D. J. et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J. Allergy Clin. Immunol. 146(203–206), e3 (2020).
Bradding, P. et al. ACE2, TMPRSS2, and furin gene expression in the airways of people with asthma-implications for COVID-19. J. Allergy Clin. Immunol. 146, 208–211 (2020).
Grifoni, A. et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181(1489–1501), e15 (2020).
Braun, J. et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 587, 270–274 (2020).
Acknowledgements
CS is supported by the Universities Giessen and Marburg Lung Center (UGMLC), the German Center for Lung Research (DZL), University Hospital Giessen and Marburg (UKGM) research funding according to article 2, section 3 cooperation agreement, and the Deutsche Forschungsgemeinschaft (DFG)-funded SFB 1021 (C04), KFO 309 (P10), and SK 317/1-1 (Project No. 428518790) as well as by the Foundation for Pathobiochemistry and Molecular Diagnostics. KCN is supported by NIH Grants U01AI140498-03 and R01AI140134-01, The Sunshine Foundation, and the Sean N. Paker Center for Allergy and Asthma Research at Stanford University.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Conception or design of the work: C.S., K.C.N.; Data collection: K.B., A.K., F.C., V.H.; Data analysis and interpretation: C.S., K.C.N., K.B., A.K., P.T., N.T., K.E., P.N.; Drafting the article: C.S., K.C.N., K.B., M.C., A.K.; Critical revision of the article: C.S., K.C.N., K.B., M.C., A.K., H.R.; Final approval of the version to be published: All.
Corresponding author
Ethics declarations
Competing interests
For CS: Consultancy and research funding, Hycor Biomedical, Bencard Allergie and Thermo Fisher Scientific; Research Funding, Mead Johnson Nutrition (MJN). For KCN: Dr. Nadeau reports Grants from National Institute of Allergy and Infectious Diseases (NIAID), Food Allergy Research & Education (FARE), End Allergies Together (EAT), Allergenis, and Ukko Pharma; Grant awardee at NIAID, National Institute of Environmental Health Sciences (NIEHS), National Heart, Lung, and Blood Institute (NHLBI), and the Environmental Protection Agency (EPA); is involved in Clinical trials with Regeneron, Genentech, AImmune Therapeutics, DBV Technologies, AnaptysBio, Adare Pharmaceuticals, and Stallergenes-Greer; Research Sponsorship by Novartis, Sanofi, Astellas, Nestle; Data and Safety Monitoring Board member at Novartis and NHLBI; Cofounded Before Brands, Alladapt, ForTra, and Iggenix; Chief Intellectual Office at FARE, Director of the World Allergy Organization (WAO) Center of Excellence at Stanford, Personal fees from Regeneron, Astrazeneca, ImmuneWorks, and Cour Pharmaceuticals; Consultant and Advisory Board Member at European Academy of Allergy and Clinical Immunology (EAACI) Research and Outreach Committee, Ukko, Before Brands, Alladapt, IgGenix, Probio, Vedanta, Centecor, Seed, Novartis, NHBLI, EPA, National Scientific Committee of Immune Tolerance Network (ITN) and NIH Programs; US patents for basophil testing, multifood immunotherapy and prevention, monoclonal antibody from plasmoblasts, and device for diagnostics. All other authors have no competing interests to declare.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Balz, K., Kaushik, A., Chen, M. et al. Homologies between SARS-CoV-2 and allergen proteins may direct T cell-mediated heterologous immune responses. Sci Rep 11, 4792 (2021). https://doi.org/10.1038/s41598-021-84320-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-84320-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.