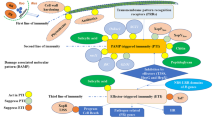
Abstract
Rhizoctonia solani is an important phytopathogenic fungus with a wide host range and worldwide distribution. The anastomosis group AG1 IA of R. solani has been identified as the predominant causal agent of rice sheath blight, one of the most devastating diseases of crop plants. As a necrotrophic pathogen, R. solani exhibits many characteristics different from biotrophic and hemi-biotrophic pathogens during co-evolutionary interaction with host plants. Various types of secondary metabolites, carbohydrate-active enzymes, secreted proteins and effectors have been revealed to be essential pathogenicity factors in R. solani. Meanwhile, reactive oxygen species, phytohormone signaling, transcription factors and many other defense-associated genes have been identified to contribute to sheath blight resistance in rice. Here, we summarize the recent advances in studies on molecular interactions between rice and R. solani. Based on knowledge of rice-R. solani interactions and sheath blight resistance QTLs, multiple effective strategies have been developed to generate rice cultivars with enhanced sheath blight resistance.
Similar content being viewed by others
Background
Rice sheath blight (RSB) caused by the necrotrophic pathogen Rhizoctonia solani Kühn is considered as one of the most devastating rice diseases worldwide (Rao et al. 2020). The disease is also called “snake skin disease”, “mosaic foot stalk”, and “rotten foot stalk” because of its special disease symptoms (Molla et al. 2020; Zhang et al. 2019b). The past decades have witnessed a sharp increase in the incidence of RSB in the field largely due to the application of high dose of nitrogen fertilizers and large-scale planting of semi-dwarf high yield cultivars (Yellareddygari et al. 2014). RSB was first reported in Japan in 1910 and subsequently spread around the world, particularly in Asia, Africa and America. In China, RSB was first reported in 1934, and it has become the second most important disease in rice at present, causing a yield loss of 10 ~ 30 % every year, even up to 50 % in the rice-growing region of Yangtze river valley and South China in epidemic years (Yu et al. 2019; Zhu et al. 2019). The annual disease area in China is about 15 ~ 20 million hm2 (Bernardes-de-Assis et al. 2009; Shu et al. 2019).
Due to the lack of resistant germplasms in rice, progress in breeding for RSB-resistant varieties is slow. At present, chemical fungicides and cultivation practices are the major approaches to preventing and managing the disease (Singh et al. 2019; Yellareddygari et al. 2014). Advances in omics, including genomics, proteomics and transcriptomics, genetic variability of R. solani, quantitative trait loci (QTL) and genetic regions for RSB resistance, and integrative management methods have been well reviewed recently (Li et al. 2019a; Molla et al. 2020). This review focuses on the molecular basis of the interactions between rice and R. solani and the molecular strategies for controlling the disease.
Overview of the Pathogens and Diseases
Thanatephorus cucumeris, the sexual stage of the causal agent of RSB, belongs to the Corticiaceae family in the Homenomycetales order of the Basidiomycetes class. The soil-borne pathogen causes diseases in many monocot and dicot plants (Anderson et al. 2017; Zhang et al. 2009). The genetically closely R. solani isolates are compatible and form a fused hyphal network involving fusion of cell wall, cytoplasm and nuclei, whereas the genetically distant isolates may not anastomose (Carling et al. 2002). Accordingly, R. solani is assigned into 14 anastomosis groups (AG1 to AG13, AGB1) with high genetic diversity based on their compatibility for hyphal fusion with known tester isolates (Carling et al. 2002; Singh et al. 2019). Based on the differences in sclerotium morphology and host plants, AG1 isolates are further divided into three subgroups, including IA, IB and IC. R. solani AG1 IA has been identified as the dominant anastomosis group causing RSB (Singh et al. 2019; Taheri and Tarighi 2011).
It is very difficult to find asexual spores of R. solani and this fungus survives in an unfavorable environment through sclerotia formed by tightly interweaving mycelia (Willetts and Bullock 1992; Sun et al. 2020b). Sclerotia are developed on infected plants late in the cropping season and are dropped into soil or water (Shu et al. 2019). Dormant sclerotia can live in the soil for several years and serve as the primary inocula for the next cropping season (Singh et al. 2019). Sclerotia can float and disperse over long distance with irrigation water and gather around rice plants on the surface of water (Yellareddygari et al. 2014; Zaeim et al. 2015). Under favorable conditions, sclerotia germinate and then the mycelia begin to grow (Feng et al. 2017; Kwon et al. 2014). R. solani gains entry into host tissues through multiple avenues, such as via infection cushions that are aggregates of complex hyphae or lobate appressoria that penetrate the cuticle, via stomata or wounds (Molle et al. 2013; Pooja and Babu 2017; Zhao et al. 2008). R. solani initially causes green-gray, water-soaked lesions on the sheath at the base of rice plants, or close to the water line in the paddy field. As the symptoms develop, the center of lesions becomes grayish white, and the edge shows dark brown. Eventually, individual lesions merge together and even cover the entire sheath. As a consequence, the sheathes become yellow and wilted, and even rot and die (Taheri and Tarighi 2011; Wang and Zheng 2018) .
Pathogenicity Factors in R. solani.
Elucidation of molecular mechanisms underlying the co-evolutionary interaction between rice and R. solani will facilitate developing novel strategies to manage RSB. Pathogen-secreted molecules play an important role in the pathogen-host interaction. It has been reported that the infection cushions are involved in enzymatic degradation and necrosis occurs in plants before mycelial invasion of R. solani (Groth and Nowick 1992; Singh et al. 2019). Besides, many defense genes in host plants are specifically down-regulated after R. solani infection (Rao et al. 2020; Mayo et al. 2015; Xia et al. 2017). These findings indicate that R. solani has evolved to secrete some substances, which actively regulate host immunity and promote virulence. Indeed, a wide arsenal of effectors, carbohydrate-active enzymes and various types of secondary metabolites secreted by R. solani have been identified to modulate host immunity (Costanzo et al. 2011; Wang et al. 2014; Wei et al. 2020).
Secreted Proteins and Effectors
It is generally believed that some effector proteins secreted by hemi-biotrophic and necrotrophic pathogens can inhibit pattern-triggered immunity (PTI) at the early and transient establishment stage of infection, while others induce cell death at the late necrotrophic stage of infection and ultimately contribute to pathogen virulence (Li et al. 2019c; Wei et al. 2020). Through comparative genomics and secretome analyses, 985 secreted proteins and 103 small cysteine-rich effector candidates were predicted in R. solani AG1 IA genome (Zheng et al. 2013). The effectors AG1IA_09161, AG1IA_05310 and AG1IA_07795 containing glycosyltransferase GT family 2, cytochrome C oxidase assembly protein CtaG/cox11 and peptidase inhibitor I9 domains, respectively, have been identified to induce cell death in rice and maize plants. Eighteen putative effectors were subsequently identified and categorized into three classes of verified effectors through the hierarchical clustering method (Zheng et al. 2013). Based on transcriptome analyses, hundreds of putative secreted protein-encoding genes and dozens of candidate effector genes are up-regulated during the infection of R. solani AG1 IA (Ghosh et al. 2018; Xia et al. 2017). Among them, a nucleoside diphosphate-linked moiety X (Nudix) domain-containing protein (AG1IA_02392), NACHT domain-containing protein (AG1IA_06487) and BTB domain-containing protein (AG1IA_03906) are preferentially expressed during R. solani infection of the susceptible rice cultivars (Rao et al. 2020). Although Nudix effectors are widely distributed in plant pathogens and probably play essential roles in pathogenesis, very few have been characterized (Dong et al. 2016). Therefore, it is important to reveal the role of AG1IA_02392 in R. solani pathogenicity (Rao et al. 2020). Recently, a putative lipase effector AGLIP1 has been demonstrated to induce cell death in Nicotiana benthamiana and rice protoplasts. Interestingly, ectopic expression of AGLIP1 suppresses basal defenses and promotes bacterial multiplication in Arabidopsis ( Li et al. 2019c). Besides, a highly conserved fungal effector RsIA_NP8 in R. solani anastomosis group AG1 is localized predominantly in the chloroplasts and triggers non-host cell death when transiently expressed in N. benthamiana (Wei et al. 2020).
Chitin, an unbranched homopolymer of 1, 4-β linked N-acetyl-D-glucosamine (GlcNAc), is the second most abundant polysaccharide on earth and is the major component of fungal cell walls. As a major pathogen-associated molecular pattern (PAMP) in rice, chitin is recognized by the pattern recognition receptor (PRR) CEBiP and coreceptor OsCERK1 on plant cell membrane, and thus initiating immune responses (Gong et al. 2020; Sanchez-Vallet et al. 2020; Shimizu et al. 2010). For successful infection, fungal pathogens have evolved to secrete certain effector proteins to interfere with PAMP recognition by PRRs. In the soil-borne Verticillium and Fusarium fungal pathogens, secretory polysaccharide deacetylase (PDA1) proteins deacetylate chitin oligomers into ligand-inactive chitosan, and thereby preventing chitin-triggered immunity and promoting virulence (Gao et al. 2019). Gene expression profiling reveals that a chitin deacetylase-encoding gene (CL8196Contig1) is significantly up-regulated during R. solani infection (Ghosh et al. 2018). Therefore, it is of interest to investigate whether R. solani exploits the same strategy to inhibit chitin-triggered host immunity. Besides, R. solani encodes the carbohydrate binding module 14 (CBM14) family of putative chitin-binding proteins similar to Cladosporum fulvum Avr4 and Ecp6, which suppress chitin-triggered immunity through blocking the release of chitin from fungal cell walls by plant chitinases and competing with CEBiP for binding of chitin oligosaccharides, respectively (Anderson et al. 2017; de Jonge et al. 2010; van den Burg et al. 2006). Additionally, the genes encoding xylanase and inhibitor I9 domain-containing proteins are transcriptionally up-regulated during R. solani infection. Both proteins can induce plant cell death when transiently expressed in N. benthamiana (Anderson et al. 2017). Collectively, the effectors in R. solani might function as important pathogenicity factors. Elegant studies from Sclerotinia sclerotiorum also substantiate the essentiality of effector proteins in virulence and pathogenicity of the necrotrophic pathogens (Liang and Rollins 2018). Next, it is interesting to identify the effectors that inhibit pattern-triggered immunity to establish initial colonization and also the effectors that induce cell death and contribute to necrotrophic stage.
Secondary Metabolites
R.solani secretes a variety of secondary metabolites, including host-selective toxins and biologically active molecules. These factors contribute to pathogen virulence through breaking host physical barriers and interfering with normal physiological functions and host defenses (Brooks 2007; Costanzo et al. 2011; Howlett 2006). The host-specific toxin in R. solani has been partially purified and identified as a carbohydrate consisting of mannose, N-acetylglucosamine, glucose and N-acetylgalactosamine. Highly virulent R. solani isolates produce more host-specific toxin than weakly virulent isolates (Chen et al. 2009; Vidhyasekaran et al. 1997; Yang et al. 2011).
Other biologically active molecules produced by R. solani include oxalic acid (OA), 3-methylthiopropionic acid (MTPA), phenylacetic acid (PAA) and its derivatives (Brooks 2007; Hu et al. 2018; Vidhyasekaran et al. 1997; Yang et al. 2014). OA produced by necrotrophic pathogens is an essential virulence factor for successful infection. The highly virulent isolates of R. solani have been identified to produce more OA than weakly virulent isolates (Nagarajkumara et al. 2005). During R. solani infection, OA inhibits the synthesis of various phenolic substances and acts synergistically with polygalacturonases (PGs) to cause pH instability and cell wall degradation (Liang et al. 2018; Qi et al. 2017; Rollins and Dickman, 2001). Besides, OA triggers programmed cell death in host plants, and thus promoting infection (Cessna et al. 2000; Heller et al. 2013; Kabbage et al. 2013). PAA treatment, similar to R. solani infection, causes severe disease symptoms on maize sheath (Cook et al. 2016; Hu et al. 2018). Besides, PAA treatment and R. solani infection have similar effects on the synthesis of secondary metabolites, including traumatin, phytosphingosine, vitexin 2’’ O-beta-D-glucoside, rutin and 2,4-dihydroxy-2H-1,4-benzoxazin-3(4H)-one (DIBOA)-glucoside, while inhibit the generation of the jasmonic acid (JA) precursors 3-oxo-2-(2’-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0) and 12-oxo-phytodienoic acid (OPDA) (Ahmad et al. 2011; Hu et al. 2018; Yang et al. 2016). The PAA metabolism pathway has been well identified and the phytotoxin inhibits seed germination and chlorophyll synthesis and functions as a virulence factor in the potato pathogen AG3 (Chen et al. 2009; Lakshman et al. 2006). Unexpectedly, it fails to directly isolate or detect PAA in R. solani AG1 IA. The amount of PAA and its derivatives detected in the culture medium of the AG1 IA isolates is also significantly lower than that from AG3 and AG4, indicating PAA might not be the primary toxin for AG1 IA (Bartz et al. 2012; Kankam et al. 2018). It has also been demonstrated that MTPA is essential for mycelial production and promotes disease progression (Kankam et al. 2018).
Furthermore, the cytochrome P450s (CYPs)-encoding genes involving in the biosynthesis of primary and secondary metabolites including phytotoxins are differentially regulated during R. solani infection and sclerotial development, implying that the CYPs and CYP-associated genes contribute to R. solani pathogenicity (Shu et al. 2019; Xia et al. 2017). These findings further highlight the essentiality of secondary metabolites in R. solani pathogenicity (Ghosh et al. 2019; Keller et al. 2005; Vidhyasekaran et al. 1997; Yang et al. 2014).
Carbohydrate‐active Enzymes
For successful invasion, plant fungal pathogens generally secrete various types of carbohydrate-active enzymes (CAZymes), particularly including cell wall-degrading enzymes (CWDEs) (Molla et al. 2020; Rao et al. 2020). CAZymes mediate the degradation of cellulose, hemicellulose and pectin in host cell walls and break down the physical barrier of plant immune system, and thus enhancing pathogen virulence (Zheng et al. 2013; Xia et al. 2017). CWDEs in R. solani are categorized into seven major families including PG, endo-β- 1,4-glucanase, polymethylgalacturonase, pectin methylesterase, filter paper enzyme, polymethylgalacturonase trans-eliminas and pectin methyltrans-eliminase (Silva et al. 2018; Xue et al. 2018; Rathinam et al. 2020). As a group of hydrolytic enzymes, PGs act on polygalacturonic acid, a key component of plant cell wall, by cleaving the α-(1,4)-glycosidic bonds, and thus causing hydrolysis of pectate. Pathogens of rice, soybean, maize and other crops have evolved multiple PGs to maximize their offensive potential (Chen et al. 2006; Gawade et al. 2017; Zhou et al. 2016). R. solani AG1 IA selectively secretes various pectinases according to the cell wall characteristics of different host species (Zheng et al. 2013). RNAseq analysis revealed that 30 genes encoding pectin-degrading enzymes in R. solani, especially PG_04727, PG_01811, PG_06500, PE_09779 and AG1IA_01129, were greatly induced during infection, indicating that these pectin-degrading enzymes are likely involved in R. solani AG1 IA infection of rice (Rao et al. 2020). Many CAZyme genes encoding pectin lyases, cellobiose dehydrogenase and β-glucanases are also up-regulated at different levels during R. solani infection of the hosts (Ghosh et al. 2018; Rao et al. 2020). Besides, endo-1,4-β-xylanase A breaks down hemicellulose in plant cell wall through decomposing linear β-1,4-xylan into xylose to facilitate pathogen penetration. The xylanase shows increased expression in highly susceptible rice plants after R. solani infection (Prathi et al. 2018).
In addition, other transcriptome analyses suggest that a variety of glycosyltransferases (GTs), glycoside hydrolases (GHs), polysaccharide lyases (PLs), carbohydrate esterases (CEs) and non-catalytic CBMs are the important CAZymes involving in the degradation of host cell walls and in R. solani virulence (Ghosh et al. 2018; Xia et al. 2017; Zheng et al. 2013). Sixteen GH-, GT-, and PL-encoding genes exhibit significantly higher expression levels in the rice susceptible cultivars compared with the resistant cultivars after R. solani inoculation, which might explain the phenotype of higher degree of necrosis in susceptible cultivars (Rao et al. 2020). Notably, much more CAZymes are up-regulated in R. solani during the necrotrophic phase compared with the colonization stage, implying that CAZymes in necrotrophic pathogens play more important roles in virulence than those in biotrophic pathogens (Blanco-Ulate et al. 2014; Ghosh et al. 2018; Zhao et al. 2013). This statement can stand close scrutiny since biotrophic pathogens generally avoid damaging the cells to acquire nutrients and restrict the cell wall degradation, which products are often recognized as damage-associated molecular patterns and thus triggering plant defenses.
Countermeasures from the Host
To counteract the effects from pathogenicity factors in plant pathogens, plants develop multiple layers of defenses against pathogen attacks. During pathogen infection, PAMPs are recognized by plant PRRs, and thus triggering PTI. As the first layer of plant defense, PTI responses include the activation of defense gene expression and mitogen-activated protein kinase (MAPK) cascades, reactive oxygen species (ROS) burst, accumulation of secondary metabolites and defense-related phytohormone signaling pathways (Bigeard et al. 2015; Yu et al. 2017; Zipfel 2014). On the other hand, intercellular immune receptors (R proteins) in host plants recognize certain pathogen effectors and therefore cause hypersensitive responses, which are rapid and robust defense responses called effector-triggered immunity (Jones and Dangl 2006; Peng et al. 2018). The two branches of plant immunity are perfectly coordinated and might cross-talk with each other to form a barrier against external biological invasion.
Reactive Oxygen Species
In general, ROS has a dual function in regulating plant defenses. ROS can function as a signal molecule initiating defense responses against fungal pathogens. In response to R. solani challenge, plants generate ROS and initiate various defense responses, such as phytoalexin biosynthesis, activation of defense-related genes and callose deposition in cell walls (Gechev et al. 2006). H2O2 activates phenylalanine ammonia lyase (PAL)-mediated biosynthesis of phenolic compounds including lignin in the shikimate pathway, and thereby improves plant antioxidant capacity and thickens cell walls (Molla et al. 2013). Particularly in the early stage of infection, plants prevent pathogen infection by enhancing cell wall lignification and producing a variety of secondary metabolites. Comprehensive KEGG enrichment analyses also indicate that phenylpropanoid and phenylalanine metabolism plays an important role in preventing infection in RSB resistant varieties (Kwon et al. 2014; Shi et al. 2020; Zhang et al. 2017).
Interestingly, the rice resistant line exhibits a significant less ROS accumulation compared with the susceptible line at 48 h post inoculation of R. solani (Oreiro et al. 2020). In this case, pathogen attack may stimulate excessive ROS accumulation in the susceptible plants and thereby inducing cell death, which is conducive to the growth and colonization of necrotrophic fungi (Heller and Tudzynski 2011). Therefore, ROS might function as a signal for R. solani to switch from the establishment to the necrotrophic stage (Noctor et al. 2018; Oreiro et al. 2020). Besides, exogenous and endogenous ROS has a promoting effect on the formation and development of R. solani sclerotia, while trehalose, a ROS-scavenger, functions as an inhibitor to sclerotial development, suggesting that trehalose might serve as a new antioxidant fungicide to inhibit sclerotial differentiation in R. solani AG-1 IA ( Wang et al. 2018).
To effectively balance the role of ROS during the interaction between rice and R. solani, many smart strategies have evolved to regulate the ROS level to reduce damage and activate defense in rice. A quantitative trait locus qLN1128 enriched in defense-related genes is involved in the activation of the genes related to the ROS-redox pathway and alleviates ROS accumulation in rice cells, and thus delaying R. solani colonization (Oreiro et al. 2020). The catalase OsCATC and chloroplast glutathione peroxidase involving in scavenging ROS are found in hub gene network in rice and might play an important role in maintaining normal ROS level in rice after R. solani infection (Alam and Ghosh 2018). In addition, previous studies revealed that various peroxidases can function to maintain the balance of H2O2 and play an important role in cell wall regeneration and thickening (Bindschedler et al. 2006; Marjamaa et al. 2009).
Phytohormones and Transcription Factors
Phytohormone signaling pathways are required for plant defenses against various pathogens. Typically, SA confers resistance against biotrophic and hemibiotrophic pathogens, whereas JA and ethylene synergistically cooperate to activate defenses against necrotrophs (Singh et al. 2018). The ethylene insensitive mutant sickle (skl) is highly susceptible to R. solani AG8, while the transgenic plants overexpressing ethylene response factors (ERFs) or the ethylene biosynthesis enzyme ACS2 exhibit resistance to R. solani (Anderson et al. 2010; Anderson et al. 2018; Helliwell et al. 2013). Ethylene-insensitive protein 2 (EIN2) is significantly up-regulated in the resistant and susceptible rice varieties during R. solani infection (Shi et al. 2020). These findings support that ethylene signaling positively regulates rice resistance to RSB. An elegant study has revealed that siR109944, a type of tourist-miniature inverted-repeat transposable element-derived small interfering RNA, suppresses rice immunity to sheath blight by affecting auxin homeostasis. The small RNA targets F-Box domain- and LRR-containing protein 55 (FBL55) encoding transport inhibitor response 1 (TIR1)-like protein. The siR109944-overexpressing rice plants have a significantly enhanced susceptibility to R. solani, while FBL55-overexpression causes the transgenic plants to be more resistant to RSB (Qiao et al. 2020). Additionally, multiple indeterminate domain (IDD) proteins including LPA1 (Loose Plant Architecture 1), IDD3 and IDD13 form a transcription factor complex and regulate expression of the auxin efflux carrier gene PIN-FORMED 1a (PIN1a) in rice through binding to its promoter (Sun et al. 2019; Sun et al. 2020a). Interestingly, LPA1 and IDD13 positively regulate PIN1a expression and RSB resistance, while IDD3 is a negative regulator in PIN1a expression and RSB resistance (Sun et al. 2019; Sun et al. 2020a). The G-protein γ subunit DEP1 (dense and erect panicle 1) interacts with LPA1 and suppresses DNA binding ability of LPA1 and thereby inhibiting expression of PIN1a. Consistently, mutation of G-protein γ subunit DEP1 can promote rice resistance to RSB (Liu et al. 2020; Sun et al. 2019; Sun et al. 2020a). Furthermore, auxin application enhances rice resistance to sheath blight (Qiao et al. 2020; Sun et al. 2019). These results indicate that auxin signaling positively regulates RSB resistance and that application of auxin analogues might effectively protect field crops against the disease.
The transcription factors including NAC, WRKY, and basic leucine zipper (bZIP) family members differentially regulate the expression of various defense genes and are involved in modulating resistance to R. solani (Olsen et al. 2005; Zhang et al. 2018). NAC transcription factors are involved in the regulation of signal transduction pathways mediated by salicylic acid (SA), JA, ethylene and abscisic acid (Zhao et al. 2020). The osnac4 mutant shows reduced necrosis symptom after pathogen infection, while OsNAC4 overexpression promotes R. solani infection in rice, indicating that OsNAC4 negatively regulates sheath blight resistance in rice (Kaneda et al. 2009). By contrast, the NAC transcription factor XNDL positively modulates RSB resistance through activating the ethylene signaling pathway (Wang 2019). Multiple WRKY family members are also important components of plant defense against pathogen infection (Gallou et al. 2012; Shi et al. 2020; Zheng et al. 2013). OsWRKY30 and OsWRKY80 are positive regulators in rice resistance against sheath blight (Peng et al. 2012; Peng et al. 2016). Silencing of OsWRKY80 in rice remarkably reduces resistance to R. solani (Peng et al., 2016). OsWRKY80 expression is induced by R. solani infection, exogenous JA and ethylene, but not by SA. OsWRKY80 elevates OsWRKY4 expression through specifically binding to the promoter of OsWRKY4 and both of transcription factors function synergistically to increase rice defenses against R. solani infection (Peng et al. 2016; Wang et al. 2015a). Over-expression of WRKY13 in rice results in significantly increased expression of WRKY12, TIFY9 and PR2 and enhances resistance to R. solani (Jimmy and Babu 2019). WRKY33 and WRKY70 are significantly induced in the rice susceptible cultivars in comparison with the resistant cultivars after R. solani infection. Various types of transcription factors are often involved in regulating different phytohormone signaling, and thereby modulating rice defenses against sheath blight. WRKY70 acts as an activator of SA-induced genes and a repressor of JA-responsive genes (Shi et al. 2020; Zhang et al. 2017). The transcription factor RAVL1 negatively regulates rice resistance to R. solani through activating the expression of brassinosteroid-related genes (Helliwell et al. 2011; Yuan et al. 2018). Paradoxically, RAVL1 also activates ethylene-mediated signaling, which positively regulates rice resistance against sheath blight (Yuan et al. 2018). These findings indicate that rice plants have evolved distinct strategies to defend against various pathogens, which might be mediated by different transcription factors.
Other Signal Molecules
As an important secondary messenger, Ca2+ regulates plant physiology and metabolism and plays an important role in plant disease resistance (Pusztahelyi et al. 2015). After R. solani infection, most of the genes involving in the calcium signaling pathway in rice are down-regulated at 12 h but increased at 24 h, which is more significant in resistant varieties compared with susceptible varieties, implying that these genes are involved in regulating sheath blight resistance (Zhang et al. 2017). The calcium-binding proteins OsBON1 and OsBON3 have been identified as negative regulators of broad-spectrum disease resistance to both bacterial and fungal pathogens including R. solani, which regulate the balance between immunity and agronomic traits in rice (Yin et al. 2018).
To combat OA action, host plants have evolved to effectively decompose oxalate by synthesizing and secreting oxalate oxidases (Karmakar et al. 2015; Molla et al. 2013). Bacteria and fungi are also able to produce oxalate decarboxylases (ODCs) to inhibit OA accumulation (Liang et al. 2015; Qi et al. 2017). During R. solani colonization of host plants, several ODC genes were significantly up-regulated (Ghosh et al. 2019; Liang et al. 2015). The ODC genes are essential for the formation of appressorium in S. sclerotiorum, another necrotrophic fungal pathogen, probably because ODCs function in OA detoxification and pH homeostasis or contribute to catabolic energy generation (Liang et al. 2015).
As a countermeasure to fungal PGs, plant polygalacturonase-inhibiting proteins (PGIPs) specifically recognize and bind to fungal PGs, and thereby inhibit the cell wall degradation activity and prevent fungal infection (Rathinam et al. 2020). The interaction of PGs and PGIPs results in the production of oligogalacturonides, which are important signals to induce PGIP gene expression and plant defense responses (Davidsson et al. 2017). The expression of the PGIP family genes is also significantly induced by R. solani infection, wounding, SA and JA (Feng et al. 2016; Zhu et al. 2019). It has been demonstrated that multiple PGIPs from different plant species except OsPGIP2 confer a wide spectrum of inhibitory activities to PGs and play an essential role in resistance against various fungal pathogens including R. solani (Wang et al. 2015b; Zhu et al. 2019).
Lessons from the Rice - R. solani Interaction
No cultivar with complete resistance has been identified after screening of thousands of rice cultivars from various rice growing regions (Li et al. 2019a; Molla et al. 2020; Shi et al. 2020). Due to the lack of resistant germplasms to RSB, successes in conventional breeding programs for RSB resistant varieties are very limited. By contrast, many pathogenicity factors in R. solani and the underlying mechanisms have been revealed in the molecular interaction between rice and R. solani (Table 1). Besides, many defense-related genes have been also identified to be associated with sheath blight resistance in rice (Table 2). The knowledge provides a solid foundation to create RSB resistant germplasms through precisely manipulating the expression of target genes via transgenic technology, gene editing and RNA interference (Table 1).
Silencing Essential Pathogenicity Genes via RNA Interference in the Fight Against R. solani
Cross-kingdom trafficking of small RNAs (sRNAs) between hosts and pathogens opens an avenue to develop disease resistant transgenic plants producing sRNAs and double-stranded RNAs, which can silence fungal pathogenicity genes (Huang et al. 2019; Wang et al. 2016). The host delivered RNA interference (HD-RNAi) technology has been developed to silence two PATHOGENICITY MAP KINASE 1 (PMK1) homologues, RPMK1-1 and RPMK1-2 in R. solani. The transgenic rice plants show an increased resistance to RSB (Tiwari et al. 2017). Besides, silencing of the key pathogenicity gene AG1IA_04727 encoding polygalacturonase via HD-RNAi significantly enhances rice resistance to R. solani (Rao et al. 2019). These results indicate that the targeting of key pathogenicity genes in R. solani by HD-RNAi is a novel and promising strategy for durable control of RSB.
Targeting Essential Pathogenicity Factors in R. solani via Transgenic Technology
The attempts at inhibiting the PG activity via PGIP overexpression are also successful in suppressing R. solani infection. Overexpression of OsPGIP1 significantly improves rice resistance to RSB (Chen et al. 2016; Rathinam et al. 2020; Wang et al. 2015b). Although OsPGIP2, a homolog of OsPGIP1, has no inhibitory activity to PGs, the mutant protein OsPGIP2L233F has been identified to have the PG inhibition activity. Overexpression of OsPGIP2L233F confers rice resistance to R. solani (Chen et al. 2019). Furthermore, the transgenic rice plants constitutively expressing ZmPGIP3 exhibit significantly elevated expression of some rice PGIP genes and enhanced resistance to sheath blight compared with the wild-type plants (Zhu et al. 2019). Importantly, these transgenic plants do not show any detrimental phenotypic or agronomic effect. The findings indicate that genome editing and natural allele mining of plant PGIP genes provide important strategies to improve RSB resistance in rice.
Since OA is an essential pathogenicity factor for necrotrophic pathogens, expression of OA-detoxifying enzymes in host plants leads to enhanced resistance against necrotrophic pathogens including R. solani (Nagarajkumara et al. 2005; Liang and Rollins 2018). Overexpression of the rice oxalate oxidase 4 gene (OsOXO4) and simultaneous overexpression of OsOXO4 and the chitinase gene OsCHI11 driven by green tissue-specific promoters both significantly confer enhanced and durable resistance to sheath blight (Karmakar et al. 2015; Molla et al. 2013). Expression of the oxalate decarboxylase Bacisubin, an oxalate-degrading enzyme from Bacillus subtilis, also enhances resistance to RSB and fungal blast diseases (Qi et al. 2017). In the transgenic plants expressing oxalate oxidases and ODCs, OA released by R. solani is degraded by these OA-detoxifying enzymes to generate H2O2. Hydrogen peroxide plays a key role in activating defense responses, such as phytoalexin biosynthetic pathways, hypersensitive response, systemic acquired resistance, and subsequently induces the expression of PR genes TS and RC24 genes (Molla et al. 2013; Qi et al. 2017).
As another essential virulence factor in R. solani, the phytotoxin PAA induces the production of the cytotoxic metabolite methylglyoxal (MG) in rice as a common consequence of many abiotic and biotic stresses. The transgenic rice plants overexpressing glyoxalase for MG detoxification have been demonstrated to have much less accumulation of MG and enhanced resistance towards damage caused by PAA. The finding provides another transgenic technology to develop RSB resistant rice plants (Gupta et al. 2017). The mechanism of observed tolerance of the glyoxalase-overexpressing plants towards diverse abiotic and biotic stresses involves enhanced detoxification and reduced oxidative damage, leading to better protection of chloroplast and mitochondrial ultrastructure and maintained photosynthetic efficiency under stress conditions (Gupta et al. 2017).
Interestingly, α-1,3-glucan produced by R. solani masks chitin to evade PRR recognition, thus promoting pathogen infection. Besides, α-1,3-glucan is essential for the maintenance of the infection-specific hyphal structure (Fujikawa et al. 2012). These findings provide a new virulence target for developing rice plants against R. solani infection. Actually, the transgenic rice plants expressing and secreting bacterial α-1,3-glucanase show strong resistance not only to R. solani but also to the phylogenetically distant ascomycete Cochlioborus miyabeanus and Magnaporthe oryzae (Fujikawa et al. 2012).
Enhancing Sheath Blight Resistance by Manipulating Expression of Plant Defense-associated Genes
Although no complete resistance gene for sheath blight has been identified in rice, many successful attempts have been performed to develop resistant rice lines by expressing defense-associated genes. Non expresser of pathogenesis-related genes 1 (NPR1) was first identified in Arabidopsis to be a master regulator of systemic acquired resistance, which confers broad-spectrum resistance to various pathogens (Fu and Dong 2013). Tissue-specific expression of Arabidopsis NPR1 gene in rice enhances sheath blight resistance without phenotypic and agronomic costs (Molla et al. 2016). The transgenic indica rice lines expressing Brassica juncea NPR1 also exhibit enhanced resistance to R. solani (Sadumpati et al. 2013). Overexpression of pathogenesis-related genes such as PR3 and PR5 results in enhanced resistance to R. solani, manifested by reduced disease lesion sizes in the transgenic rice plants as compared with the wild-type rice plants (Datta et al. 1999; Datta et al. 2001; Datta et al. 2002). In addition, over-expression of OsGSTU5, a tau class glutathione-S-transferase, in rice effectively increases the activities of superoxide dismutase and peroxidase, and thereby reduces accumulation of H2O2 and O2−, and enhances rice resistance to sheath blight (Tiwari et al. 2020). Various MAPKs play important roles in plant adaptive responses to biotic stresses. Silencing of OsMAPK20-5 remarkably reduces resistance of rice to M. oryzae, but enhances resistance to R. solani (Liu et al. 2019). Another group of small, highly basic, cysteine-rich antimicrobial peptides called defensins from various plant species exhibit strong antimicrobial activities. Expression of such proteins as Ace-AMP1 from Allium cepa, Dm-AMP1 from Dahlia merckii and Rs-AFP2 from Raphanus sativus in rice greatly suppresses the growth of M. oryzae and R. solani (Jha et al. 2009; Jha and Chattoo 2010; Patkar and Chattoo 2006). ASR2 (short for abscisic acid, stress and ripening 2) specifically binds to an important cis-element GT-1 and activates the expression of the rice defense-related gene Os2H16. Over-expression of the stress-responsive gene OsASR2 enhances resistance against Xanthomonas oryzae pv. oryzae and R. solani, and tolerance to drought in rice (Li et al. 2018). Collectively, many defense-associated genes can be manipulated to improve sheath blight resistance in rice (Table 2).
QTL for Disease Resistance to RSB
It is well recognized that rice resistance to sheath blight is a quantitative trait controlled by multiple genes (Zuo et al. 2014a). Therefore, identification, mapping and subsequent characterization of RSB resistance QTL will be of great significance for sheath blight resistance breeding in rice (Jia et al. 2012; Molla et al. 2020; Taguchi-Shiobara et al. 2013; Yadav et al. 2015).
Since the first RSB resistance QTL was identified in 1995, more than 110 RSB resistance QTLs have been mapped to different chromosomes in rice (Molla et al. 2020; Wen et al. 2015; Zhang et al. 2019a). However, only qSBR9-2, qSBR11 − 1, qSB-9TQ and qSB-11LE have been finely mapped and no RSB resistance QTL has yet been isolated in rice (Channamallikarjuna et al. 2010; Zuo et al. 2013; Zuo et al. 2014a). A total of 14, 12, 12 and 26 putative genes have been predicted in the qShB9-2, qSB-9TQ, qSB-11LE and qSBR11-1 regions, respectively (Channamallikarjuna et al. 2010; Zhang et al. 2019a; Zuo et al. 2013; Zuo et al. 2014b). The SBR-9 locus in a 12.8-Mbp region between the markers Nag08KK18184 and Nag08KK18871 contains at least two putative resistance genes (Liu et al. 2009; Taguchi-Shiobara et al. 2013). In addition, a novel chitinase gene LOC_Os11g47510 in the QTL qSBR11-1 region of the RSB resistant rice cultivar Tetep has been functionally verified. Expression of the chitinase gene in the RSB susceptible rice line significantly inhibits the growth and branching of R. solani hyphae and enhances rice resistance to RSB (Richa et al. 2017). A major-effect QTL for durable resistance containing a cluster of 12 germin-like protein (OsGLP) genes has been identified on chromosome 8 in rice. The more the OsGLP genes in this gene cluster are silenced by RNAi, the more the transgenic lines are susceptible to R. solani. The disease resistance conferred by OsGLPs at the QTL region, which is highly conserved in the grass family, is broad-spectrum (Manosalva et al. 2009). More importantly, an F-box protein ZmFBL41 has recently been identified to confer resistance to banded leaf and sheath blight in maize through a genome-wide association study (Li et al. 2019b). The isolation of the maize sheath blight resistance gene offers a promising avenue to generate sheath blight resistant cultivars in different crop plants.
Multiple QTLs for RSB resistance have been detected in several resistant varieties, including Teqing, Jasmine 85, Zhaiyeqing 8, Xiangzaoxian 19, Tetep and Pecos (Datta et al. 2001). Some major QTLs for RSB resistance, such as qSB-9TQ, qSB-11LE and qSB-11HJX, have been utilized in resistance breeding program. Pyramiding disease resistance QTLs has been considered as an important strategy to develop RSB resistant cultivars. The lines carrying qSB-11LE and qSB-11HJX have a significantly lower disease level than the recurrent parent and the lines with single QTL under the same genetic background, indicating that QTL pyramiding can further increase the resistance to sheath blight (Li et al. 2019a). It has been also demonstrated that pyramiding of two QTLs (qSB9-2 and qSB12-1) and three QTLs (qSB7TQ, qSB9TQ and qSB11LE) has higher levels of resistance than a single QTL (Wang et al. 2012; Yin et al. 2008). Therefore, simultaneous introduction of multiple resistance alleles and stacking of multiple QTLs in susceptible varieties increase rice resistance to RSB (Chen et al. 2014; Pinson et al. 2008; Zuo et al. 2008).
Sheath blight resistance QTLs are often mapped to the same region as the QTLs for heading date and plant height (Pinson et al. 2005; Sharma et al. 2009). For instance, four QTLs for RSB resistance have been identified in F8 recombinant inbred lines of two rice varieties from the United States, which are allelic to the QTLs for plant height and heading date (Goad et al. 2020). The flag leaf angle, length and plant compactness are also tightly linked with the RSB resistance QTLs, qRlh11, qSBR11-3, qSBR11-1, qSBR9-1, qShB3-2 and qSB-9 (Hossain et al. 2016). The reason for the correlation between morphological traits and resistance may be related to canopy microenvironment. Rice canopy density has a great impact on humidity and temperature, which are the important environment factors for pathogen infection and multiplication.
Conclusions
Sheath blight has become one of the most important diseases in rice. Genetic resistance has been well considered as the most economic, effective and environmentally friendly strategy to control crop diseases. However, no RSB resistance gene has been isolated and identified. Increasing knowledge on molecular interaction between R. solani and host plants reveals many pathogenicity factors in R. solani and defense-associated genes in rice. Accordingly, the effective strategies to develop RSB resistant germplasms include disarming essential pathogenicity factors in R. solani via host-derived RNAi and transgenic technology, manipulating expression of plant defense-associated genes, and pyramiding the RSB resistance QTLs.
Availability of Data and Materials
Not applicable.
References
Alam NB, Ghosh A (2018) Comprehensive analysis and transcript profiling of Arabidopsis thaliana and Oryza sativa catalase gene family suggests their specific roles in development and stress responses. Plant Physiol Biochem 123:54–64
Ahmad S, Veyrat N, Gordon-Weeks R, Zhang Y, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, Ton J (2011) Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol 157:317–327
Anderson JP, Lichtenzveig J, Gleason C, Oliver RP, Singh KB (2010) The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with Rhizobia. Plant Physiol 154:861–873
Anderson JP, Sperschneider J, Win J, Kidd B, Yoshida K, Hane J, Saunders DGO, Singh KB (2017) Comparative secretome analysis of Rhizoctonia solani isolates with different host ranges reveals unique secretomes and cell death inducing effectors. Sci Rep 7:10410
Anderson JP, Kidd BN, Garg G, Singh KB (2018) Transcriptome analysis reveals class IX ethylene response factors show specific up-regulation in resistant but not susceptible Medicago truncatula lines following infection with Rhizoctonia solani. Eur J Plant Pathol 152:549–554
Baisakh N, Datta K, Oliva N, Ona I, Rao G, Mew T, Datta SK (2001) Rapid development of homozygous transgenic rice using anther culture harboring rice chitinase gene for enhanced sheath blight resistance. Plant Biotechnol 18:101–108
Bartz FE, Glassbrook NJ, Danehower DA, Cubeta MA (2012) Elucidating the role of the phenylacetic acid metabolic complex in the pathogenic activity of Rhizoctonia solani anastomosis group 3. Mycologia 104:793–803
Bernardes-de-Assis J, Storari M, Zala M, Wang WX, Jiang DH, Dong LS, Jin MS, McDonald BA, Ceresini PC (2009) Genetic structure of populations of the rice-infecting pathogen Rhizoctonia solani AG-1 IA from China. Phytopathology 99:1090–1099
Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8:521–539
Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, Plotnikov J, Denoux C, Hayes T, Gerrish C, Davies DR, Ausubel FM, Bolwell GP (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47:851–863
Blanco-Ulate B, Morales-Cruz A, Amrine KCH, Labavitch JM, Powell ALT, Cantu D (2014) Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front Plant Sci 5:435
Brooks SA (2007) Sensitivity to a phytotoxin from Rhizoctonia solani correlates with sheath blight susceptibility in rice. Phytopathology 97:1207–1212
Carling DE, Baird RE, Gitaitis RD, Brainard KA, Kuninaga S (2002) Characterization of AG-13, a newly reported anastomosis group of Rhizoctonia solani. Phytopathology 92:893–899
Cessna SG, Sears VE, Dickman MB, Low PS (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12:2191–2199
Channamallikarjuna V, Sonah H, Prasad M, Rao GJN, Chand S, Upreti HC, Sharma TR (2010) Identification of major quantitative trait loci qSBR11-1 for sheath blight resistance in rice. Mol Breeding 25:155–166
Chen XJ, Zhang H, Xu JY, Tong YH, Ji ZL (2006) Cell wall degrading enzymes produced by Rhizoctonia solani and their pathogenicity to rice plants. Jiangsu J Agr Sci 22:24–28
Chen XJ, Xi Y, Tong YH, Meng LJ, Ji ZL, Xu JY (2009) Pathogenic mechanism of phytotoxin produced by Rhizoctonia solani, the causal pathogen of rice sheath blight. Acta Phytopathol Sin 39:439–443
Chen ZX, Zhang YF, Feng F, Feng MH, Jiang W, Ma YY, Pan CH, Hua HL, Li GS, Pan CB, Zuo SM (2014) Improvement of japonica rice resistance to sheath blight by pyramiding qSB-9TQ and qSB-7TQ. Field Crop Res 161:118–127
Chen XJ, Chen Y, Zhang LN, Xu B, Zhang JH, Chen ZX, Tong YH, Zuo SM, Xu JY (2016) Overexpression of OsPGIP1 enhances rice resistance to sheath blight. Plant Dis 100:388–395
Chen XJ, Chen YW, Zhang L, He Z, Huang BL, Chen C, Zhang QX, Zuo SM (2019) Amino acid substitutions in a polygalacturonase inhibiting protein (OsPGIP2) increases sheath blight resistance in rice. Rice 12:56
Cook SD, Nichols DS, Smith J, Chourey PS, McAdam EL, Quittenden L, Quittenden L, Ross JJ (2016) Auxin biosynthesis: Are the indole-3-acetic acid and phenylacetic acid biosynthesis pathways mirror images? Plant Physiol 171:1230–1241
Costanzo S, Jackson AK, Brooks SA (2011) High-resolution mapping of Rsn1, a locus controlling sensitivity of rice to a necrosis-inducing phytotoxin from Rhizoctonia solani AG1-IA. Theor Appl Genet 123:33–41
Datta K, Velazhahan R, Oliva N, Ona I, Mew T, Khush GS, Muthukrishnan S, Datta SK (1999) Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor Appl Genet 98:1138–1145
Datta K, Tu J, Oliva N, Ona II, Velazhahan R, Mew TW, Muthukrishnan S, Datta SK (2001) Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Sci 160:405–414
Datta K, Baisakh N, Maung T, Thet KM, Tu J, Datta S (2002) Pyramiding transgenes for multiple resistance in rice against bacterial blight, yellow stem borer and sheath blight. Theor Appl Genet 106:1–8
Davidsson P, Broberg M, Kariola T, Sipari N, Pirhonen M, Palva ET (2017) Short oligogalacturonides induce pathogen resistance-associated gene expression in Arabidopsis thaliana. BMC Plant Biol 17:19
de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MHAJ, Thomma BPHJ (2010) Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329:953–955
Dong SM, Wang YC (2016) Nudix effectors: A common weapon in the arsenal of plant pathogens. PLoS Pathog 12:e1005704
Feng CS, ZhangX, Wu T, Yuan B, Ding XH, Yao FY, Chu ZH (2016) The polygalacturonase-inhibiting protein 4 (OsPGIP4), a potential component of the qBlsr5a locus, confers resistance to bacterial leaf streak in rice. Planta 243:1297–1308
Feng SJ, Shu CW, Wang CJZ, Jiang SF, Zhou EX (2017) Survival of Rhizoctonia solani AG-1 IA, the causal agent of rice sheath blight, under different environmental conditions. J Phytopathol 165:44–52
Fu ZQ, Dong XN (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64:839–863
Fujikawa T, Sakaguchi A, Nishizawa Y, Kouzai Y, Minami E, Yano S, Koga H, Meshi T, Nishimura M (2012) Surface alpha-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog 8:e1002882
Gallou A, Declerck S, Cranenbrouck S (2012) Transcriptional regulation of defence genes and involvement of the WRKY transcription factor in arbuscular mycorrhizal potato root colonization. Funct Integr Genomic 12:83–198
Gao F, Zhang BS, Zhao JH, Huang JF, Jia PS, Wang S, Zhang J, Zhou JM, Guo HS (2019) Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat Plants 5:1167–1176
Gawade DB, Perane RR, Suryawanshi AP, Deokar CD (2017) Extracellular enzymes activity determining the virulence of Rhizoctonia bataticola, causing root rot in soybean. Physiol Mol Plant Pathol 100:49–56
Gechev TS, Breusegem FV, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28:1091–1101
Ghosh S, Kanwar P, Jha G (2018) Identification of candidate pathogenicity determinants of Rhizoctonia solani AG1-IA, which causes sheath blight disease in rice. Curr Genet 64:729–740
Ghosh S, Mirza N, Kanwar P, Tyagi K, Jha G (2019) Genome analysis provides insight about pathogenesis of Indian strains of Rhizoctonia solani in rice. Funct Integr Genomic 19:799–810
Goad DM, Jia YL, Gibbons A, Liu Y, Gealy D, Caicedo AL, Olsen KM (2020) Identification of novel QTL conferring sheath blight resistance in two weedy rice mapping populations. Rice 13:21
Gong BQ, Wang FZ, Li JF (2020) Hide-and-seek: chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci 25:805–816
Groth DE, Nowick EM (1992) Selection for resistance to rice sheath blight through number of infection cushions and lesion type. Plant Dis 76:721–723
Gupta BK, Sahoo KK, Ghosh A, Tripathi AK, Anwar K, Das P, Singh AK, Pareek A, Sopory SK, Singla-Pareek SL (2017) Manipulation of glyoxalase pathway confers tolerance to multiple stresses in rice. Plant Cell Environ 41:1186–1200
Heller J, Tudzynski P (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu Rev Phytopathol 49:369–390
Heller A, Witt-Geiges T (2013) Oxalic acid has an additional, detoxifying function in Sclerotinia sclerotiorum pathogenesis. PLoS One 8:e72292
Helliwell EE, Wang Q, Yang YN (2011) Transgenic rice with inducible overproduction of ethylene exhibits broad-spectrum disease resistance. Phytopathology 101:S71
Helliwell EE, Wang Q, Yang YN (2013) Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol J 11:33–42
Hossain MK, Jena KK, Bhuiyan MAR, Wickneswari R (2016) Association between QTLs and morphological traits toward sheath blight resistance in rice (Oryza sativa L). Breeding Sci 66:613–626
Howlett BJ (2006) Secondary metabolite toxins and nutrition of plant pathogenic fungi. Curr Opin Plant Biol 9:371–375
Hu WJ, Pan XL, Li FF, Dong WB (2018) UPLC-QTOF-MS metabolomics analysis revealed the contributions of metabolites to the pathogenesis of Rhizoctonia solani strain AG-1-IA. PLoS One 13:e0192486
Huang CY, Wang H, Hu P, Hamby R, Jin HL (2019) Small RNAs-big players in plant-microbe interactions. Cell Host Microbe 26:173–182
Jha S, Tank HG, Prasad BD, Chattoo BB (2009) Expression of Dm-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani. Transgenic Res 18:59–69
Jha S, Chattoo BB (2010) Expression of a plant defensin in rice confers resistance to fungal phytopathogens. Transgenic Res 19:373–384
Jia L, Yan W, Zhu C, Agrama HA, Jackson A, Yeater K, Li XB, Huang BH, Hu BL, McClung A, Wu D (2012) Allelic analysis of sheath blight resistance with association mapping in rice. PLoS One 7:e32703
Jimmy LJ, Babu S (2019) Gene network mediated by WRKY13 to regulate resistance against sheath infecting fungi in rice (Oryza sativa L). Plant Sci 280:269–282
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kabbage M, Williams B, Dickman MB (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog 9:e1003287
Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che FS (2009) The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J 28:926–936
Kankam F, Sowley ENK, Qiu HZ (2018) Influence of 3-methylthiopropionic acid (MTPA) produced by Rhizoctonia solani AG-3 on yield and dry matter accumulation of potato. Asian J Res Crop Sci 2:1–9
Karmakar S, Molla KA, Chanda PK, Sarkar SN, Datta SK, Datta K (2015) Green tissue-specific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta 243:115–130
Karmakar S, Molla KA, Das K, Sarkar SN, Datta SK, Datta K (2017) Dual gene expression cassette is superior than single gene cassette for enhancing sheath blight tolerance in transgenic rice. Sci Rep 7:7900
Keller NP, Turner G, Bennett JW (2005) Fungal secondary metabolism - From biochemistry to genomics. Nat Rev Microbiol 3:937–947
Kumar KK, Poovannan K, Nandakumar R, Thamilarasi K, Geetha C, Jayashree N, Kokiladevi E, Raja JAJ, Samiyappan Ramasamy, Sudhakar D, Balasubramanian P (2003) A high throughput functional expression assay system for a defence gene conferring transgenic resistance on rice against the sheath blight pathogen, Rhizoctonia solani. Plant Sci 165:969–976
Kwon YS, Kim SG, Chung WS, Bae H, Jeong SW, Shin SC, Jeong MJ, Park SC, Kwak YS, Bae DW, Lee YB (2014) Proteomic analysis of Rhizoctonia solani AG-1 sclerotia maturation. Fungal Biol 118:433–443
Lakshman DK, Liu CY, Mishra PK, Tavantzis S (2006) Characterization of the arom gene in Rhizoctonia solani, and transcription patterns under stable and induced hypovirulence conditions. Curr Genet 49:166–177
Li N, Wei ST, Chen J, Yang FF, Kong LG, Chen CX, Ding XH, Chu ZH (2018) OsASR2 regulates the expression of a defence-related gene, Os2H16, by targeting the GT-1 cis-element. Plant Biotechnol J 16:771–783
Li MY, Wang JN, Wang GD, Feng ZM, Ye YH, Jiang W, Zuo T, Zhang YF, Chen XJ, Pan XB, Ma YY, Chen ZX, Zuo SM (2019a) Improvement of japonica rice resistance to sheath blight disease by incorporating quantative resistance genes qSB-11HJX and qSB-9TQ. J Yangzhou Univ 40:1–7
Li N, Lin B, Wang H, Li XM, Yang FF, Ding XH, Yan JB, Chu ZH (2019b) Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat Genet 51:1540–1548
Li S, Peng XW, Wang YL, Hua KY, Xing F, Zheng YY, Liu W, Sun WX, Wei SH (2019c) The effector AGLIP1 in Rhizoctonia solani AG1 IA triggers cell death in plants and promotes disease development through inhibiting PAMP-triggered immunity in Arabidopsis thaliana. Front Microbiol 10:2228
Liang XF, Moomaw EW, Rollins JA (2015) Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol Plant Pathol 16:825–836
Liang XF, Rollins JA (2018) Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 108:1128–1140
Lin W, Anuratha C, Datta K, Potrykus I, Muthukrishnan S, Datta SK (1995) Genetic engineering of rice for resistance to sheath blight. Nat Biotechnol 13:686–691
Liu G, Jia Y, Correa-Victoria FJ, Prado GA, Yeater KM, McClung A, Correll JC (2009) Mapping quantitative trait loci responsible for resistance to sheath blight in rice. Phytopathology 99:1078–1084
Liu XL, Li JC, Noman A, Lou YG (2019) Silencing OsMAPK20-5 has different effects on rice pests in the field. Plant Signal Behav 14:e1640562
Liu JM, Mei Q, Xue CY, Wang ZY, Li DP, Zhang YX, Xuan YH (2020) Mutation of G-protein γ subunit DEP1 increases planting density and resistance to sheath blight disease in rice. Plant Biotechnol J. doi:https://doi.org/10.1111/pbi.13500
Manosalva PM, Davidson RM, Liu B, Zhu X, Hulbert SH, Leung H, Leach JE (2009) A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol 149:286–296
Marjamaa K, Kukkola EM, Fagerstedt KV (2009) The role of xylem class III peroxidases in lignification. J Exp Bot 60:367–376
Mayo S, Gutierrez S, Malmierca MG, Lorenzanal A, Campelol MP, Hermosa R, Casquero PA (2015) Influence of Rhizoctonia solani and Trichoderma spp. in growth of bean (Phaseolus vulgaris L) and in the induction of plant defense-related genes. Front Plant Sci 6:685
Molla KA, Karmakar S, Chanda PK, Ghosh S, Sarkar SN, Datta SK, Datta K (2013) Rice oxalate oxidase gene driven by green tissue-specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Mol Plant Pathol 14:910–922
Molla KA, Karmakar S, Chanda PK, Sarkar SN, Datta SK, Datta K (2016) Tissue-specific expression of Arabidopsis NPR1 gene in rice for sheath blight resistance without compromising phenotypic cost. Plant Sci 250:105–114
Molla KA, Karmakar S, Molla J, Bajaj P, Varshney RK, Datta SK, Datta K (2020) Understanding sheath blight resistance in rice: the road behind and the road ahead. Plant Biotechnol J 18:895–915
Nagarajkumara M, Jayaraj J, Muthukrishnan S, Bhaskaran R, Velazhahan R (2005) Detoxification of oxalic acid by Pseudomonas fluorescens strain PfMDU2: implications for the biological control of rice sheath blight caused by Rhizoctonia solani. Microbiol Res 160:291–298
Noctor G, Reichheld JP, Foyer CH (2018) ROS-related redox regulation and signaling in plants. Sem Cell Dev Biol 80:3–12
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Oreiro EG, Grimares EK, Grande G, Quibod IL, Roman-Reyna V, Oliva R (2020) Genome-wide associations and transcriptional profiling reveal ROS regulation as one underlying mechanism of sheath blight resistance in rice. Mol Plant-Microbe Interact 33:212–222
Patkar RN, Chattoo BB (2006) Transgenic indica rice expressing ns-LTP-like protein shows enhanced resistance to both fungal and bacterial pathogens. Mol Breeding 17:159–171
Peng XX, Hu YJ, Tang XK, Zhou PL, Deng XB, Wang HH, Guo ZJ (2012) Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236:1485–1498
Peng XX, Wang HH, Jang JC, Xiao T, He HH, Jiang D, Tang XK (2016) OsWRKY80-OsWRKY4 module as a positive regulatory circuit in rice resistance against. Rhizoctonia solani Rice 9:63
Peng YJ, Wersch RV, Zhang YL (2018) Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol Plant Microbe Interact 31:403–409
Pinson SRM, Capdevielle FM, Oard JH (2005) Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci 45:503–510
Pinson SRM, Oard JH, Groth D, Miller R, Marchetti MA, Shank AR, Jia MH, Jia Y, Fjellstrom RG, Li Z (2008) Registration of TIL:455, TIL:514, and TIL:642, three rice germplasm lines containing introgressed sheath blight resistance alleles. J Plant Regist 2:251–254
Pooja S, Babu S (2017) Responses of rice to Rhizoctonia solani and its toxic metabolite in relation to expression of Osmyb4 transcription factor. Plant Protect Sci 53:208–215
Prathi NB, Palit P, Madhu P, Ramesh M, Laha GS, Balachandran SM, Madhav MS, Sundaram RM, Mangrauthia SK (2018) Proteomic and transcriptomic approaches to identify resistance and susceptibility related proteins in contrasting rice genotypes infected with fungal pathogen Rhizoctonia solani. Plant Physiol Biochem 130:258–266
Pusztahelyi T, Holb IJ, Pocsi I (2015) Secondary metabolites in fungus-plant interactions. Front Plant Sci 6:573
Qiao LL, Zheng LY, Sheng C, Zhao HW, Jin HL, Niu DD (2020) Rice siR109944 suppresses plant immunity to sheath blight and impacts multiple agronomic traits by affecting auxin homeostasis. Plant J 102:948–964
Qi ZQ, Yu JJ, Shen LR, Yu ZC, Yu MN, Du Y, Zhang RS, Song TQ, Yin XL, Zhou YX, Li HH, Wei Q, Liu YF (2017) Enhanced resistance to rice blast and sheath blight in rice (Oryza sativa L) by expressing the oxalate decarboxylase protein Bacisubin from Bacillus subtilis. Plant Sci 265:51–60
Rao TB, Chopperla R, Methre R, Punniakotti E, Venkatesh V, Sailaja B, Reddy MR, Yugander A, Laha GS, Madhav MS, Sundaram RM, Ladhalakshmi D, Balachandran SM, Mangrauthia SK (2019) Pectin induced transcriptome of a Rhizoctonia solani strain causing sheath blight disease in rice reveals insights on key genes and RNAi machinery for development of pathogen derived resistance. Plant Mol Biol 100:59–71
Rao TB, Chopperla R, Prathi NB, Balakrishnan M, Prakasam V, Laha GS, Balachandran SM, Mangrauthia SK (2020) A comprehensive gene expression profile of pectin degradation enzymes reveals the molecular events during cell wall degradation and pathogenesis of rice sheath blight pathogen Rhizoctonia solani AG1-IA. J Fungi 6:71
Rathinam M, Rao U, Sreevathsa R (2020) Novel biotechnological strategies to combat biotic stresses: polygalacturonase inhibitor (PGIP) proteins as a promising comprehensive option. Appl Microbiol Biotech 104:2333–2342
Richa K, Tiwari IM, Devanna BN, Botella JR, Sharma V, Sharma TR (2017) Novel chitinase gene LOC_Os11g47510 from Indica rice Tetep provides enhanced resistance against sheath blight pathogen Rhizoctonia solani in rice. Front Plant Sci 8:596
Rollins JA, Dickman MB (2001) PH signaling in Sclerotinia sclerotiorum: identification of a pacC/RIM1 homolog. Appl Environ Microbiol 67:75–81
Sadumpati V, Kalambur M, Vudem DR, Kirti PB, Khareedu VR (2013) Transgenic indica rice lines, expressing Brassica juncea nonexpressor of pathogenesis-related gene 1 (BjNPR1), exhibit enhanced resistance to major pathogens. J Biotechnol 166:114–121
Sanchez-Vallet A, Tian H, Rodriguez-Moreno L, Valkenburg DJ, Saleem-Batcha R, Wawra S, Kombrink A, Verhage L, Jonge R, Esse HPeter, Zuccaro A, Croll D, Mesters JR, Thomma BPHJ (2020) A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PLoS Pathog 16:e1008652
Sharma A, Mcclung AM, Pinson SRM, Kepiro JL, Shank AR, Tabien RE, Fjellstrom R (2009) Genetic mapping of sheath blight resistance QTLs within tropical japonica rice cultivars. Crop Sci 49:256–264
Shi W, Zhao SL, Liu K, Sun YB, Ni ZB, Zhang GY, Tang HS, Zhu JW, Wan BJ, Sun HQ, Dai JY, Sun MF, Yan GH, Wang AM, Zhu GY (2020) Comparison of leaf transcriptome in response to Rhizoctonia solani infection between resistant and susceptible rice cultivars. BMC Genom 21:245
Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, Shibuya N (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64:204–214
Shu CW, Zhao M, Anderson JP, Garg G, Singh KB, Zheng WB, Wang CJ, Yang M, Zhou EX (2019) Transcriptome analysis reveals molecular mechanisms of sclerotial development in the rice sheath blight pathogen Rhizoctonia solani AG1-IA. Funct Integr Genomic 19:743–758
Silva MS, Arraes FBM, Campos MA, Grossi-de-Sa M, Fernandez D, Candido ES, Cardoso MH, Franco OL, Grossi-de-Sa MF (2018) Review: Potential biotechnological assets related to plant immunity modulation applicable in engineering disease-resistant crops. Plant Sci 270:72–84
Singh PK, Nag A, Arya P, Kapoor R, Singh A, Jaswal R, Sharma TR (2018) Prospects of understanding the molecular biology of disease resistance in rice. Int J Mol Sci 19:1141
Singh P, Mazumdar P, Harikrishna JA, Babu S (2019) Sheath blight of rice: a review and identification of priorities for future research. Planta 250:1387–1407
Sridevi G, Sabapathi N, Meena P, Nandakumar R, Samiyappan R, Muthukrishnan S, Veluthambi K (2003) Transgenic indica rice variety Pusa Basmati 1 constitutively expressing a rice chitinase gene exhibits enhanced resistance to Rhizoctonia solani. J Plant Biochem Biotech 12:93–101
Sridevi G, Parameswari C, Sabapathi N, Raghupathy V, Veluthambi K (2008) Combined expression of chitinase and beta-1,3-glucanase genes in indica rice (Oryza sativa L.) enhances resistance against Rhizoctonia solani. Plant Sci 175:283–290
Sripriya R, Raghupathy V, Veluthambi K (2008) Generation of selectable marker-free sheath blight resistant transgenic rice plants by effificient co-transformation of a cointegrate vector T-DNA and a binary vector T-DNA in one Agrobacterium tumefaciens strain. Plant Cell Rep 27:1635–1644
Sun Q, Li TY, Li DD, Wang ZY, Li S, Li DP, Han X, Liu JM, Xuan YH (2019) Overexpression of Loose Plant Architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN-FORMED 1a in rice. Plant Biotechnol J 17:855–857
Sun Q, Li DD, Chu J, Yuan DP, Li S, Zhong LJ, Han X, Xuan YH (2020a) Indeterminate domain proteins regulate rice defense to sheath blight disease. Rice 13:15
Sun XY, Liu DM, Wang YY, Ma AM (2020b) Biogenesis of macrofungal sclerotia: influencing factors and molecular mechanisms. Appl Microbiol Biotechnol 104:4227–4234
Taguchi-Shiobara F, Ozaki H, Sato H, Maeda H, Kojima Y, Ebitani T, Yano M (2013) Mapping and validation of QTLs for rice sheath blight resistance. Breed Sci 63:301–308
Taheri P, Tarighi S (2011) Cytomolecular aspects of rice sheath blight caused by Rhizoctonia solani. Eur J Plant Pathol 129:511–528
Tiwari IM, Jesuraj A, Kamboj R, Devanna BN, Botella JR, Sharma TR (2017) Host delivered RNAi, an efficient approach to increase rice resistance to sheath blight pathogen (Rhizoctonia solani). Sci Rep 7:7521
Tiwari M, Srivastava S, Singh PC, Mishra AK, Chakrabarty D (2020) Functional characterization of tau class glutathione-S-transferase in rice to provide tolerance against sheath blight disease. 3 Biotech 10:84
van den Burg HA, Harrison SJ, Joosten M, Vervoort J, de Wit P (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19:1420–1430
Vidhyasekaran P, Ponmalar TR, Samiyappan R, Velazhahan R, Vimala R, Ramanathan A, Paranidharan V, Muthukrishnan S (1997) Host-specific toxin production by Rhizoctonia solani, the rice sheath blight pathogen. Phytopathology 87:1258–1263
Wang Y, Pinson SRM, Fjellstrom RG, Tabien RE (2012) Phenotypic gain from introgression of two QTL, qSB9-2 and qSB12-1, for rice sheath blight resistance. Mol Breeding 30:293–303
Wang XL, Jiang N, Liu JL, Liu WD, Wang GL (2014) The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence 5:722–732
Wang HH, Meng J, Peng XX, Tang XK, Zhou PL, Xiang JH, Deng XB (2015a) Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol Biol 89:157–171
Wang R, Lu LX, Pan XB, Hu ZL, Ling F, Yan Y, Liu YM, Lin Y (2015b) Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol Biol 87:181–191
Wang M, Weiberg A, Lin FM, Thomma BP, Huang HD, Jin H (2016) Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants 2:1–10
Wang AJ, Zheng AP (2018) Characteristics and control measures of rice sheath blight. Chinese Rice 24:124–126
Wang CJZ, Pi L, Jiang SF, Yang M, Shu CW, Zhou EX (2018) ROS and trehalose regulate sclerotial development in Rhizoctonia solani AG-1 IA. Fungal Biol 122:322–332
Wang ZY (2019) Mechanism research of transcription factor XNDLs regulates rice sheath blight resistance. Master’s Thesis
Wei MM, Wang AJ, Liu Y, Ma L, Niu XY, Zheng AP (2020) Identification of the novel effector RsIA_NP8 in Rhizoctonia solani AG1 IA that induces cell death and triggers defense responses in non-host plants. Front Microbiol 11:1115
Wen ZH, Zeng YX, Ji ZJ, Yang CD (2015) Mapping quantitative trait loci for sheath blight disease resistance in Yangdao 4 rice. Genet Mol Res 14:1636–1649
Willetts HJ, Bullock S (1992) Developmental biology of sclerotia. Mycol Res 96:801–816
Xia Y, Fei BH, He JY, Zhou ML, Zhang DH, Pan LX, Li SC, Liang YY, Wang LX, Zhu JQ, Li P, Zheng AP (2017) Transcriptome analysis reveals the host selection fitness mechanisms of the Rhizoctonia solani AG1 IA pathogen. Sci Rep 7:10120
Xue X, Cao ZX, Zhang XT, Wang Y, Zhang YF, Chen ZX, Pan XB, Zuo SM (2016) Overexpression of OsOSM1 enhances resistance to rice sheath blight. Plant Dis 100:1634–1642
Xue CY, Zhou RJ, Li YJ, Xiao D, Fu JF (2018) Cell-wall-degrading enzymes produced in vitro and in vivo by Rhizoctonia solani, the causative fungus of peanut sheath blight. Peer J 6:e5580
Yadav S, Anuradha G, Kumar RR, Vemireddy LR, Sudhakar R, Donempudi K, Venkata D, Jabeen F, Narasimhan YK, Marathi B, Siddiq EA (2015) Identification of QTLs and possible candidate genes conferring sheath blight resistance in rice (Oryza sativa L.). SpringerPlus 4:175
Yang YQ, Yang M, Li MH, Jia Y, Zhang DT, Zhou EX (2011) Isolation and characterization of a phytotoxin from Rhizoctonia solani: the causal agent of rice sheath blight. Asian J Chem 23:3500–3508
Yang YQ, Yang M, Lan B, Zhou EX, Li XM (2014) Research progress in pathogenic mechanism of rice sheath blight pathogen. Chin Agr Sci Bulletin 30:245–250
Yang W, Xu XN, Li Y, Wang YZ, Li M, Wang Y, Ding XH, Chu ZH, Lam HM (2016) Rutin-mediated priming of plant resistance to three bacterial pathogens initiating the early SA signal pathway. PLoS One 11:e0146910
Yellareddygari SKR, Reddy MS, Kloepper JW, Lawrence KS, Fadamiro H (2014) Rice sheath blight: a review of disease and pathogen management approaches. J Plant Pathol Microb 5:4
Yin YJ, Zuo SM, Wang H, Chen ZX, Ma YY, Zhang YF, Gu SL, Pan XB (2008) Pyramiding effects of three quantitative trait loci for resistance to sheath blight using near-isogenic lines of rice. Chin J Rice Sci 22:340–346
Yin X, Zou BH, Hong XX, Gao MJ, Yang WB, Zhong XB, He Y, Kuai P, Lou YG, Huang JR, Hua J, He ZH (2018) Rice copine genes OsBON1 and OsBON3 function as suppressors of broad-spectrum disease resistance. Plant Biotechnol J 16:1476–1487
Yu X, Feng BM, He P, Shan LB (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55:109–137
Yuan P, Zhang C, Wang ZY, Zhu XF, Xuan YH (2018) RAVL1 activates brassinosteroids and ethylene signaling to modulate response to sheath blight disease in rice. Phytopathology 108:1104–1113
Yu YD, Sun HJ, Xia ZH (2019) Progress on biological control of rice sheath blight. Mol Plant Breeding 17:600–605
Zaeim AN, Amirabad NM, Drakhshan A, Noorozi M, Moradi K (2015) A review for rice sheath blight disease. IJSRST 1:233–237
Zhang CQ, Liu YH, Ma XY, Feng Z, Ma ZH (2009) Characterization of sensitivity of Rhizoctonia solani, causing rice sheath blight, to mepronil and boscalid. Crop Prot 28:381–386
Zhang JF, Chen L, Fu CL, Wang LX, Liu HN, ChengYZ, Li SC, Deng QM, Wang SQ, Zhu J, Liang YY, Li P, Zheng AP (2017) Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Front Plant Sci 8:1422
Zhang JF, Zhao WJ, Fu R, Fu CL, Wang LX, Liu HN, Li SC, Deng QM, Wang SQ, Zhu J, Liang YY, Li P, Zheng AP (2018) Comparison of gene co-networks reveals the molecular mechanisms of the rice (Oryza sativa L.) response to Rhizoctonia solani AG1 IA infection. Funct Integ Genomic 18:545–557
Zhang F, Zeng D, Zhang CS, Lu JL, Chen TJ, Xie JP, Zhou YL (2019a) Genome-wide association analysis of the genetic basis for sheath blight resistance in rice. Rice 12:93
Zhang SW, Yang Y, Li KT (2019b) Occurrence and control against rice sheath blight. Biol Dis Sci 42:87–91
Zhao CJ, Wang AR, Shi YJ, Wang LQ, Liu WD, Wang ZH, Lu GD (2008) Identification of defense-related genes in rice responding to challenge by Rhizoctonia solani. Theor Appl Genet 116:501–516
Zhao ZT, Liu HQ, Wang CF, Xu JR (2013) Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom 14:274
Zhao CM, Huang XQ, Yin FY, Li DQ, Chen Y, Chen L, Cheng ZQ (2020) Research progress on NAC transcription factor family in Oryza sativa L. Plant Sci J 38:278–287
Zheng AP, Lin RM, Zhang DH, Qin PG, Xu LZ, Ai P, Ding L, Wang YR, Chen Y, Liu Y, Sun ZG, Feng HT, Liang XX, Fu RT, Tang CQ, Li Q, Zhang J, Xie ZL, Deng QM, Li SC, Wang S, Zhu J, Wang LX, Liu HN, Li P (2013) The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat Commun 4:1424
Zhu G, Liang EX, Lan X, Li Q, Qian JJ, Tao HX, Zhang MJ, Xiao N, Zuo SM, Chen JM, Gao Y (2019) ZmPGIP3 gene encodes a polygalacturonase-inhibiting protein that enhances resistance to sheath blight in rice. Phytopathology 109:1732–1740
Zhou SY, Liu Y, Zhang M, Li BD, Chen XJ, Liang WX (2016) Comparison of the virulence and cognate virulence factors of multinucleate, binucleate and uninucleate Rhizoctonia isolates, causing sheath blight on maize plants. Eur J Plant Pathol 145:501–506
Zipfel C (2014) Plant pattern-recognition receptors. Trends Immunol 35:345–351
Zuo SM, Zhang L, Wang H, Yin YJ, Zhang YF, Chen ZX, Ma YY, Pan X (2008) Prospect of the QTL-qSB-9Tq utilized in molecular breeding program of japonica rice against sheath blight. J Genet Genomics 35:499–505
Zuo SM, Yin YJ, Pan CH, Chen ZX, Zhang YF, Gu SL, Zhu LH, Pan XB (2013) Fine mapping of qSB-11LE, the QTL that confers partial resistance to rice sheath blight. Theor Appl Genet 126:1257–1272
Zuo SM, Zhang YF, Yin YJ, Li GZ, Zhang GW, Wang H, Chen ZX, Pan XB (2014a) Fine-mapping of qSB-9TQ, a gene conferring major quantitative resistance to rice sheath blight. Mol Breeding 34:2191–2203
Zuo SM, Zhu YJ, Yin YJ, Wang H, Zhang YF, Chen ZX, Gu SL, Pan XB (2014b) Comparison and confirmation of quantitative trait loci conferring partial resistance to rice sheath blight on chromosome 9. Plant Dis 98:957–964
Acknowledgements
Not applicable.
Funding
The work is supported by National Natural Science Foundation of China grants U19A2027 and 31630064, the Natural Science Foundation of Beijing, China (6181001) to W. S., and National Natural Science Foundation of China grants 31901956 to S. L.
Author information
Authors and Affiliations
Contributions
DL, SL and WS proposed the concept; DL, SL, SW and WS drafted the manuscript; DL, SL and WS revised and finalized the manuscript. All the authors have participated in the work sufficiently. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, D., Li, S., Wei, S. et al. Strategies to Manage Rice Sheath Blight: Lessons from Interactions between Rice and Rhizoctonia solani. Rice 14, 21 (2021). https://doi.org/10.1186/s12284-021-00466-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-021-00466-z