Abstract
Decoded neurofeedback (DecNef) is a form of closed-loop functional magnetic resonance imaging (fMRI) combined with machine learning approaches, which holds some promises for clinical applications. Yet, currently only a few research groups have had the opportunity to run such experiments; furthermore, there is no existing public dataset for scientists to analyse and investigate some of the factors enabling the manipulation of brain dynamics. We release here the data from published DecNef studies, consisting of 5 separate fMRI datasets, each with multiple sessions recorded per participant. For each participant the data consists of a session that was used in the main experiment to train the machine learning decoder, and several (from 3 to 10) closed-loop fMRI neural reinforcement sessions. The large dataset, currently comprising more than 60 participants, will be useful to the fMRI community at large and to researchers trying to understand the mechanisms underlying non-invasive modulation of brain dynamics. Finally, the data collection size will increase over time as data from newly run DecNef studies will be added.
Measurement(s) | functional brain measurement |
Technology Type(s) | 3 T MRI scanner • functional magnetic resonance imaging • Neurofeedback • machine learning |
Factor Type(s) | type of task performed: visual, preference, perceptual, or memory task |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.13578008
Similar content being viewed by others
Background & Summary
Neurofeedback, based on both functional magnetic resonance imaging (fMRI) and non-fMRI techniques, has seen a dramatic increase in the number of published studies in the last decade1. Decoded neurofeedback (DecNef), a form of closed-loop fMRI neurofeedback combined with machine learning approaches, is a more fine-grained rendering of the long-held goal of manipulating brain dynamics or representations2,3. As opposed to univariate approaches, where one measures the overall activity level within a region-of-interest (ROI), by treating each voxel in isolation, multivoxel pattern analysis (MVPA)4,5 is based on algorithms that learn to decode information distributed in patterns of activity. Because DecNef leverages MVPA, rather than using univariate approaches, it has high target specificity1,6,7. Furthermore, while participants know that a neurofeedback experiment is taking place, they are unaware of the content and purpose of the manipulation - reducing confounds due to cognitive processes, or knowledge about the manipulated dimension7,8. In its most recent declination, the experimenter can even infer the target neural representation indirectly from surrogate participants through a method called hyperalignment9,10. With such a functional alignment approach, patterns of neural activity across participants are used to construct a common, high-dimensional space through a set of linear transformations. These transformations are effectively parameters, and can be used to bring any new data patterns to/from an individual’s brain coordinate system and the model space coordinates9.
These aspects make DecNef an attractive tool to develop novel clinical applications, particularly in the context of neuropsychiatric disorders7,11,12,13,14,15. Besides clinically-oriented studies, DecNef can be used as an important paradigm in systems and cognitive neuroscience to study basic functions of the brain10,16. Several DecNef experiments have been completed to date, targeting representations and cognitive or psychological processes at different levels of the cortical hierarchy: vision and perceptual learning in early visual cortex3,17, subjective preference in cingulate cortex18, as well as perceptual confidence in a frontoparietal network19,20.
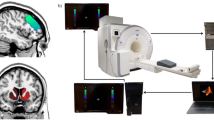
Briefly, the goal of each study contained in this data collection was to target and non-invasively modulate a specific representation in the brain. Notwithstanding that each experiment probed a different cognitive process or mental representation, all studies used the same basic design logic (Fig. 1a): (1) an initial session to acquire fMRI data that was used to train the machine learning algorithm - the MVPA or decoder construction session; 2) subsequent neurofeedback sessions, ranging from 2 to 5 days, depending on the study (described in detail in the “Data Records” section). In the decoder construction session, participants performed a simple visual (Study 2, 3), preference (Study 1), perceptual (Study 4), or memory task (Study 5), depending on the study target, while in the neurofeedback sessions the procedure was nearly identical in all cases (Fig. 1b). During neurofeedback training, participants were instructed to modulate or manipulate their brain activity in order to maximize the size of a feedback disc shown on the screen towards the end of each trial. The size of the disc reflected the amount of reward obtained on that single trial (maximum JP¥300), which was then summed to a terminal reward. Participants were told that the goal of the task was to maximize the amount of reward (bonus of up to JP¥3000, on top of JP¥8000). Unbeknownst to them, what really determined the size of the feedback disc - and therefore the amount of reward - was the likelihood that their current brain state corresponded to a target state. That is, the previously trained decoder was used in real time to infer how likely it was that a target activity pattern had occurred.
Schematic overview of the experimental design(s). (a) Each study had a first fMRI session dedicated to acquire the data necessary to construct a ‘decoder’, a machine learning classifier of brain activity patterns. (b) All studies shared the same basic trial design: a first period of 6 seconds called ‘induction’, during which participants were instructed to “modulate, or change their brain activity in order to maximize the feedback received later in the trial”. Unbeknownst to participants, brain activity patterns were fed to the previously trained decoder to calculate the likelihood that the current brain state corresponded to the target state. The likelihood was fed back to participants in the form of a disc within a circle: the larger the disc, the larger the obtained reward. The circle indicated the maximum attainable reward. Beyond this basic structure, the design was slightly modified to meet the requirement of each individual study. In study 1, a neutral face was presented for 0.5 second before the onset of the induction period. Note that in the original study actual face images were used, not cartoons. In study 2 and 3, instead of a simple visual cue indicating the induction period, participants were presented with an achromatic grating. Lastly, in study 4, a random dot kinetogram with 0% coherence was shown for 2 seconds at variable onsets during the induction period.
As of yet, only a few research groups worldwide have had the opportunity to run such technically challenging experiments. We have published elsewhere a description on how to run decoded neurofeedback experiments10. Nevertheless, a key aspect of DecNef (and neurofeedback in general) remains rather elusive: what is the underlying neural mechanism? Recent work has begun to explore this question through experiments, meta-analyses, computational models, and neural network simulations6,21,22,23,24,25,26. Reinforcement learning has been suggested as one plausible mechanism6,22, but the field would considerably benefit from having access to a rich dataset, comprising more than 60 individuals and totalling more than 200 hours of fMRI scanning time under DecNef training. This data collection consists of 5 independent studies that shared the same overall experimental design. Table 1 summarises all studies, indicating the relevant publication(s), and the main target of the neurofeedback process training.
Finally, it is important to highlight the future growth potential of the DecNef data collection. Besides data generated through our own collaborative efforts, we have recently released software to perform DecNef experiments10,27, under agreement terms that all data shall be made available to the wider scientific community through the DecNef data collection. As such, this cross-country scientific effort will result in new datasets being added semi-automatically to this data sharing project.
Methods
Ethics statement
All participants in all studies provided written informed consent. All recruitment procedures and experimental protocols were approved by the institutional review board at the Advanced Telecommunications Research Institute International (ATR, Kyoto, Japan), (approval numbers: 14–121, 12–120, 15–181, 14–140, and 16–181), and conducted in accordance with the Declaration of Helsinki.
Participants
Table 2 summarizes the demographic characteristics of participants included in each study.
Source data
Technical details about scanning parameters used in all studies are given below, and those that differed between studies, in Table 3.
Participants were scanned in 3 T MRI scanners (see Table 3 for details) with a head coil (see Table 3 for details, e.g., number of channels) at the ATR Brain Activation Imaging Center. In all experiments, 33 contiguous slices were acquired (TR = 2000 ms, voxel size = 3 × 3 × 3.5 mm3, field-of-view = 192 × 192 mm, matrix size = 64 × 64, slice thickness = 3.5 mm, 0 mm slice gap, flip angle = 80 deg) with interleaved order (e.g., [1 3 5…, 2 4 6…]), oriented parallel to the AC-PC plane, covering the entire brain. T1-weighted MR images were also obtained (magnetization-prepared rapid gradient-echo or MP-RAGE; 256 slices, TR = 2250 ms, TE = 3.06 ms, voxel size = 1 × 1 × 1 mm3, field-of-view = 256 × 256 mm, matrix size = 256 × 256, slice thickness = 1 mm, 0 mm slice gap, TI = 900 ms, flip angle = 9 deg).
All data were converted from raw DICOM images to anonymized Nifti files, the standard format in neuroimaging (Neuroimaging Informatics Technology Initiative), with the software dcm2niix28. The Nifti files from this data collection are provided as such.
Data Records
The data collection can be accessed from our institutional repository “DecNef Project Brain Data Repository” (https://bicr-resource.atr.jp/drmd/), or from the Synapse data repository (https://doi.org/10.7303/syn23530650)29. See “Usage Notes” for a detailed explanation on how to get access to, and download the data.
The data is organized according to the structure illustrated in Fig. 2. Briefly, for each study, the top folder contains a folder for each participant (e.g., “sub-01”). Within it there are three subfolders, “anat” with the raw Nifti files related to structural/anatomy scans, “func” - which is further subdivided into session-specific folders (e.g., “ses-0”: decoder, “ses-1”: first session of neurofeedback, etc.) containing all compressed Nifti files from the functional scans. Each of these subfolders additionally include the reference functional image used for re-alignment and the motion parameters computed offline. The third folder “pattern” contains preprocessed data related to the targeted brain region(s) from the decoder construction session and from the neurofeedback sessions. Preprocessed data include trial-by-trial multivoxel activity patterns corresponding to the voxels selected by the decoder algorithm (for both decoder and neurofeedback sessions)); the weights assigned to each voxel by the decoder algorithm and used in the real-time classification to infer the likelihood; the induction performance for neurofeedback sessions (i.e., the trial-by-trial likelihood of decoding the target activity pattern). Finally, the folder also contains the coordinates in native and native-TBV space of the voxels used by the decoder to compute the neurofeedback scores. The file “README.txt”, included in each study folder, provides details on the data content. The file “Participants.csv” provides demographics information on participants, as well as, where relevant, information on the training group to which participants were assigned (e.g., Study 1: increase or decrease of preference to neutral faces). The number of participants, number of neurofeedback sessions, and the target of training are summarised in Table 4.
Technical Validation
Data anonymization procedure
High resolution anatomical scans were defaced to ensure proper anonymization of structural data. Images were bias-corrected with the Statistical Parametric Mapping (SPM) Toolbox (https://www.fil.ion.ucl.ac.uk/spm/) version 12, and defacing was performed with the automated defacing tools from the FreeSurfer suite30 (see Fig. 3 for the resulting image in an example participant).
Real-time correction of head motion
Given the fine-grained, high spatial resolution requirements of the DecNef approach, a crucial aspect of a successful study pertains to a technical detail. The functional alignment of brain images used for online feedback computations need to have a very high between-sessions consistency. That is, the images have to be aligned to the original decoder construction and the alignment has to be precise at the (sub)voxel level. Even small head movements easily break this prerequisite, leading to imperfect decoding and feedback score calculations. To ensure this requirement, all studies monitored in real time the alignment between the incoming functional brain images and a template from the original decoder construction. Although Turbo BrainVoyager (TBV, Brain Innovation) was used to correct for head motion in real-time - and the displacement could be viewed by the experimenter throughout, there is no guarantee that the corrected images will be meaningful in terms of decoding, especially if there was a sudden, marked displacement.
Therefore, during real-time neurofeedback experiments, the following processing steps were applied to the raw DICOM images. First, measured functional images during the induction period underwent three-dimensional motion correction using TBV. Second, time-courses of signal intensities were extracted from each of the voxels identified by the decoder, and were shifted by 6 seconds to account for the hemodynamic delay. Third, a linear trend was removed from the time course, and the signal time course was z-score normalized for each voxel using signal intensities measured from 10 s after the onset of each fMRI run. Fourth, the data samples to calculate the feedback score were created by averaging the BOLD signal intensities of each voxel over the induction period (i.e., 3 TRs).
Functional alignment of multivoxel patterns between decoder construction and DecNef sessions
An efficient way to control for data quality in terms of the target voxels’ activation pattern is to first compute their mean activation (with data from the initial decoder construction session); then, in the real-time sessions, calculate the trial-by-trial correlation between the mean pattern and the incoming activity pattern. This approach ensures that upon significant changes in the voxels’ response patterns due to, e.g., head or body motion, a decrease of the correlation would rapidly occur and thus can be detected. At optimal levels the correlation should be within the interval r ∈[0.85 1.00], or Fisher transformed z ∈[1.26 Inf]. We confirmed that this was the case in all studies (Fig. 4), with, on average, less than 2% of the trials having z < 1.26 (Study 1: 0.13%, Study 2: 3.17%, Study 3: 0.91%, Study 4: 0.36%, Study 5: 3.74%).
Fisher transformed correlation between mean activity patterns and real-time activity patterns. High values indicate better functional alignment between the real-time measured pattern and decoder construction pattern. Each dot represents the correlation value for one trial. For each violin plot there are N (days) × M (participants) × L (runs) × J (trials) dots. The white circle at the centre of the plots represents the median, the thick grey line, the interquartile range, and the thin grey line the adjacent values. Data points with z < 1.10 were removed from the plot.
Pattern correlation measure and head motion
We next established the relationship between head motion and the pattern correlation measure. Head motion parameters were computed with SPM12, yielding 3 parameters for translation and 3 parameters for rotation. For the purpose of this analysis, we computed the mean absolute rotation and mean absolute translation by averaging the relevant 3 parameters, over the 3 TRs used in the neurofeedback experiment to compute the decoder likelihood and the pattern correlation. These two indexes of head motion (in mm) are plotted against the Fisher transformed correlation coefficients (i.e., the pattern correlation measure) pooled across all studies (Fig. 5). For statistical analysis, the single trial data were concatenated and analysed with linear mixed effects (LME) models (in Wilkinson notation, specified as y ~ 1 + m + (1 | st) + (1 + st | prt); where y: pattern correlation, 1: intercept, m: motion parameter, st: study, prt: participant). Specifically, these LME models were designed such that motion was treated as a fixed effect, study as a random effect and covariate, and individual participants as random effects nested within studies. Each model was fitted on the corresponding motion parameter (see Tables 5 and 6, respectively). The significant couplings with parameter values ≪0 for the fixed effect ‘motion’ indicate that pattern correlation was negatively influenced by head motion. Interestingly, pattern correlation appeared more sensitive to rotation movements.
Pattern correlation vs head motion. Head motion was computed as rotation in the 3D directions (a) or as translation in the 3D directions (b), and plotted against Fisher transformed pattern correlation coefficients. Data points from different studies are plotted with different colours. Each data point represents a trial from a given participant, run, and session. Statistical significance was assessed with linear mixed effects models (Tables 5, 6).
Additional sources of noise
Finally, other sources of physiological noise (e.g., heartbeat, or respiration) will also impact the multivoxel activity patterns used for real-time decoding. These sources were not directly measured in the present studies, and therefore we may only speculate on their influence. If their effects were relatively homogenous across all voxels, we expect little to no impact on the pattern correlation measure because the relationship between voxels would remain essentially unchanged. In the presence of uneven effects across voxels, we would see the pattern correlation affected similarly as with head motion. One may worry that in the first case we would not be able to detect noise distortions of the data, thus potentially invalidating the veracity of the target likelihood fed back to participants. But because all voxels’ activities first underwent baseline normalization, and then the feedback likelihood was calculated as the dot product between the voxels’ activity pattern and a weight vector, what matters is the pattern of voxels (the ‘difference’ between voxels activities). It should be noted, therefore, that due to the specificity of MVPA, it is unlikely that additional sources of noise would have significantly affected the information content of voxels’ activity patterns without also affecting the pattern correlation measure.
Usage Notes
The data is available for the purposes of scientific research, technology development, and education under the auspices of an academic, research, government or commercial entity, and is stored at https://bicr-resource.atr.jp/drmd/. Applicants should first read the privacy statement of the data collection, fill the registration form and submit their application through the website linked above. Upon clearance, each dataset will be downloadable separately. The DecNefPro consortium (https://bicr.atr.jp/decnefpro/), where the data collection is hosted, is formally recognized as a national database by the Japan Agency for Medical Research and Development (AMED), an official Agency of the Japanese Government.
Additionally, the data collection can also be accessed under the same terms at Synapse (https://doi.org/10.7303/syn23530650).
Users of the DecNef data collection are encouraged to formally cite this publication.
Code availability
To convert DICOM images to Nifti files, the open sources software dcm2niix is freely available at https://github.com/rordenlab/dcm2niix.
To construct machine learning decoders, all studies employed the freely available Sparse Logistic Regression Toolbox, implemented in Matlab. The toolbox implements classification with sparsity prior and is available for both binary and multiclass problems. The toolbox can be found at https://bicr.atr.jp/~oyamashi/SLR_WEB.html.
Software to run DecNef experiments can be obtained freely upon reception of a signed agreement form, and participating the resulting data towards the DecNef data collection. The usage of the software is intended for academic purposes. The software is implemented in Matlab and can be found at https://bicr.atr.jp/decnefpro/software/.
References
Watanabe, T., Sasaki, Y., Shibata, K. & Kawato, M. Advances in fMRI real-time neurofeedback. Trends Cogn. Sci. 21, 997–1010 (2017).
LaConte, S. M., Peltier, S. J. & Hu, X. P. Real-time fMRI using brain-state classification. Hum. Brain Mapp. 28, 1033–1044 (2007).
Shibata, K., Watanabe, T., Sasaki, Y. & Kawato, M. Perceptual learning incepted by decoded fMRI neurofeedback without stimulus presentation. Science 334, 1413–1415 (2011).
Kamitani, Y. & Tong, F. Decoding the visual and subjective contents of the human brain. Nat. Neurosci. 8, 679–685 (2005).
Norman, K., Polyn, S., Detre, G. & Haxby, J. Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends Cogn. Sci. 10, 424–430 (2006).
Shibata, K. et al. Toward a comprehensive understanding of the neural mechanisms of decoded neurofeedback. NeuroImage 188, 539–556 (2019).
Lubianiker, N. et al. Process-based framework for precise neuromodulation. Nat. Hum. Behav. 3, 436–445 (2019).
Muñoz-Moldes, S. & Cleeremans, A. Delineating implicit and explicit processes in neurofeedback learning. Neurosci. Biobehav. Rev. 118, 681–688 (2020).
Haxby, J. et al. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron 72, (2011).
Taschereau-Dumouchel, V., Cortese, A., Lau, H. & Kawato, M. Conducting decoded neurofeedback studies. Soc. Cogn. Affect. Neurosci. nsaao63 (2020).
Taschereau-Dumouchel, V. et al. Towards an unconscious neural reinforcement intervention for common fears. Proc. National Acad. Sci. 115, 201721572 (2018).
Koizumi, A. et al. Fear reduction without fear through reinforcement of neural activity that bypasses conscious exposure. Nat. Hum. Behav. 1, 0006 (2016).
Chiba, T. et al. Current status of neurofeedback for post-traumatic stress disorder: a systematic review and the possibility of decoded neurofeedback. Front. Hum. Neurosci. 13, 233 (2019).
Yamada, T. et al. Resting-state functional connectivity-based biomarkers and functional MRI-based neurofeedback for psychiatric disorders: a challenge for developing theranostic biomarkers. Int. J. Neuropsychopharmacol. 20, yx059 (2017).
Taschereau-Dumouchel, V., Kawato, M. & Lau, H. Multivoxel pattern analysis reveals dissociations between subjective fear and its physiological correlates. Mol. Psychiatry 25, 2342–2354 (2020).
Cortese, A., De Martino, B. & Kawato, M. The neural and cognitive architecture for learning from a small sample. Curr. Opin. Neurobiol. 55, 133–141 (2019).
Amano, K., Shibata, K., Kawato, M., Sasaki, Y. & Watanabe, T. Learning to associate orientation with color in early visual areas by associative decoded fMRI neurofeedback. Curr. Biol. 26, 1861–1866 (2016).
Shibata, K., Watanabe, T., Kawato, M. & Sasaki, Y. Differential activation patterns in the same brain region led to opposite emotional states. PLoS Biol. 14, e1002546 (2016).
Cortese, A., Amano, K., Koizumi, A., Kawato, M. & Lau, H. Multivoxel neurofeedback selectively modulates confidence without changing perceptual performance. Nat. Commun. 7, 13669 (2016).
Cortese, A., Amano, K., Koizumi, A., Lau, H. & Kawato, M. Decoded fMRI neurofeedback can induce bidirectional confidence changes within single participants. NeuroImage 149, 323–337 (2017).
Skottnik, L., Sorger, B., Kamp, T., Linden, D. & Goebel, R. Success and failure of controlling the real-time functional magnetic resonance imaging neurofeedback signal are reflected in the striatum. Brain Behav. e01240 (2019).
Oblak, E. F., Lewis-Peacock, J. A. & Sulzer, J. S. Self-regulation strategy, feedback timing and hemodynamic properties modulate learning in a simulated fMRI neurofeedback environment. PLoS Comput. Biol. 13, e1005681 (2017).
Emmert, K. et al. Meta-analysis of real-time fMRI neurofeedback studies using individual participant data: How is brain regulation mediated? NeuroImage 124, 806–812 (2016).
Oblak, E. F., Sulzer, J. S. & Lewis-Peacock, J. A. A simulation-based approach to improve decoded neurofeedback performance. NeuroImage 195, 300–310 (2019).
Haugg, A. et al. Can we predict real-time fMRI neurofeedback learning success from pretraining brain activity? Hum. Brain Mapp. 41, 3839–3854 (2020).
Sepulveda, P. et al. How feedback, motor imagery, and reward influence brain self-regulation using real-time fMRI. Hum. Brain Mapp. 37, 3153–3171 (2016).
Yoshioka, T. Source code for: Toolbox to run Decoded Neurofeedback (DecNef) or Functional Connectivity Neurofeedback (FCNef) experiments. https://bicr.atr.jp/decnefpro/software/ (2020).
Li, X., Morgan, P. S., Ashburner, J., Smith, J. & Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 264, 47–56 (2016).
Cortese, A. et al. DecNef Realtime MRI Dataset. Synapse https://doi.org/10.7303/syn23530650 (2020).
Bischoff-Grethe, A. et al. A technique for the deidentification of structural brain MR images. Hum. Brain Mapp. 28, 892–903 (2007).
Acknowledgements
We thank Yasuhiro Shimada for help in preparing fMRI scanning protocols tables and text, Kaori Nakamura for help in retrieving participants’ demographics, and acquiring retrospective consent from participants in studies 3 and 5, and Nobuyuki Izumihara for technical assistance in defacing and database preparation. Funded in parts by JST ERATO (Grant JPMJER1801), AMED (Grant JP17dm0107044, JP20dm0307008) to A.C. and M.K.; the National Institute of Neurological Disorders and Stroke and the National Institutes of Health (Grant R01NS088628), the National Institute of Mental Health (Grant R61MH113772) to H.L.; JSPS Kakenhi (Grant 19H01041) to K.S., the National Eye Institute (Grants R21EY028329, R01EY027841, R01EY019466) to Y.S. and T.W.; AMED (Grant JP20dm0307004, JP20dm03070) to S.C.T.
Author information
Authors and Affiliations
Contributions
A.C., K.A., A.K., K.S., V.T.D. collected the data; A.C. and S.C.T. formatted the data; A.C. performed analyses and drafted the manuscript; all authors contributed to the original design of the experiments, and revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
There is a potential financial conflict of interest; authors K.S., Y.S., T.W. and M.K. are the inventors of patents related to the neurofeedback method used to generate the data described here, and the original assignee of the patents is ATR with which several authors are affiliated. The remaining authors declare no competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Cortese, A., Tanaka, S.C., Amano, K. et al. The DecNef collection, fMRI data from closed-loop decoded neurofeedback experiments. Sci Data 8, 65 (2021). https://doi.org/10.1038/s41597-021-00845-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00845-7
This article is cited by
-
From internal models toward metacognitive AI
Biological Cybernetics (2021)







