Impact of Plastic Debris on the Gut Microbiota of Caretta caretta From Northwestern Adriatic Sea
- 1Unit of Microbiome Science and Biotechnology, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy
- 2Fano Marine Center, The Inter-Institute Center for Research on Marine Biodiversity, Resources and Biotechnologies, Fano, Italy
- 3Fondazione Cetacea Onlus, Riccione, Italy
- 4Animal and Environmental Physiology Laboratory, Department of Biological, Geological and Environmental Sciences (BiGeA), University of Bologna, Ravenna, Italy
Plastic pollution is nowadays a relevant threat for the ecological balance in marine ecosystems. Small plastic debris (PD) can enter food webs through various marine organisms, with possible consequences on their physiology and health. The loggerhead sea turtle (Caretta caretta), widespread across the whole Mediterranean Sea, is a “flagship species,” useful as indicator of the general pollution level of marine ecosystems. Ingested PD accumulate in the final section of turtles’ digestive tract before excretion. During their transit and accumulation, PD also interact with the residing microbial community, with possible feedback consequences on the host’s health. To explore the possible relationship between fecal microbial composition and PD ingestion, we collected fecal samples from 45 turtles rescued between 2017 and 2019 in the Northwestern Adriatic Sea (Italy), assessing occurrence and content of PD in the samples and in parallel the microbiome structure by 16S rRNA gene sequencing. According to our findings, almost all samples contained PD, mirroring the high level of plastic pollution in the area. We identified phylotypes associated to a high amount of PD, namely Cetobacterium somerae and other taxa, possibly responding to contamination by plastic-associated chemicals. Furthermore, putative marine pathogens were found associated to higher plastic contamination, supporting the hypothesis that PD can act as a carrier for environmental pathogenic bacteria into marine organisms. Besides confirming the role of the sea turtle as relevant flagship species for plastic pollution of the marine environment, our study paves the way to the exploration of the impact that PD ingestion can have on the microbial counterpart of large marine organisms, with potential feedback consequences on the animal and ecosystem health.
Introduction
With a global mass production of more than 350 million tons per year (PlasticsEurope, 2019), plastic is nowadays one of the major emerging pollutant in marine environments (UNEP, 2014). Indeed, it has been estimated that more than 10 million tons of plastic enter the oceans every year (Jambeck et al., 2015), becoming responsible for more than 80% of the total marine litter (European Parliament, 2019). Such amount of litter especially harms species that are not able to discriminate marine litter or confuse plastic debris (PD) with preys (Barnes et al., 2009; Schuyler et al., 2014a, b). Up to now, it has been estimated that around 260 species, including marine mammals, birds and sea turtles, are threatened by PD, via entanglement and/or ingestion (Caron et al., 2018; Isangedighi et al., 2018). Moreover, breaking down into smaller fragments and filaments (Rocha-Santos and Duarte, 2015; Peng et al., 2017), plastics enter the marine food webs through ingestion by a large number of fish and shellfish species (Barboza et al., 2018), progressively accumulating across the food chain up to the top predators, including humans (Rochman et al., 2015; Nelms et al., 2018; D’Souza et al., 2020).
Sea turtles are among the wildlife groups most impacted by plastics (Gall and Thompson, 2015), with more than half of the total individuals worldwide predicted to ingest plastics during their lifetime (Schuyler et al., 2016), making these large vertebrates important flagship species for plastic pollution of the marine environment (Foti et al., 2009). Once plastics are ingested by sea turtles, either actively (i.e., by mistaking plastic residues for pray), indirectly (feeding on animals which previously ingested plastics), or accidentally (Schuyler et al., 2016; Nelms et al., 2018), they mainly accumulate within the gastrointestinal tract, due to the inability of the animal to regurgitate items (Matiddi et al., 2017; Wilcox et al., 2018; Santos et al., 2020). Several studies have already investigated the presence of plastic pollution in different species of sea turtles, but the vast majority of them were performed on gastric contents taken from dead animals (Caron et al., 2018; Duncan et al., 2019; Digka et al., 2020; López-Martínez et al., 2020). Besides leaving a considerable gap of knowledge in monitoring the actual plastic contamination of live animals, it has been pointed out that the sole observation of the upper part of the intestinal tract, taken during necroscopy, can underestimate the real magnitude of plastic ingestion by the animal (Bjorndal et al., 1994; Pham et al., 2017). Indeed, the amount of PD is reported to increase progressively from the esophagus to the stomach and the intestine, and that debris can remain in the last part of the gut for more than 40 days before being defecated (Hoarau et al., 2014), gradually accumulating if the ingestion is not sporadic. This highlights the need to include feces as target samples in studies focused on the plastic ingestion by sea turtles (Pham et al., 2017).
The PD accumulation in the gastrointestinal tract is reported to exert sub-lethal effects on the sea turtle health, i.e., reproduction and endocrine systems dysfunctions, gastrointestinal blockage, injuries, reduced feeding, and absorption of toxic compounds (reviewed in Franzellitti et al., 2019). However, scarce knowledge is available on the mechanisms that cause those effects, calling for further investigation.
Dwelling in the last part of the intestinal tract, PD can interact with the residing microbial community, possibly influencing its compositional structure and functional properties. Gut microbiota, i.e., the bacterial community inhabiting the gastrointestinal tract of all vertebrates, is well known to play a crucial role in maintaining host physiobiological homeostasis and health, being important for food digestion, metabolism regulation, immune system functionality, and defense against pathogens’ colonization (Hooper et al., 2012; Semova et al., 2012; Godon et al., 2016). The gut microbiome is known to change its composition in relation to the presence of environmental pollutants (Evariste et al., 2019) but it has also been proposed as a site for selection of metabolic function related to the detoxification of chemical pollutants, e.g., heavy metals and pesticides, that enter the gastrointestinal tract from the environment (Itoh et al., 2018; Duan et al., 2020). Indeed, bacteria recovered from the gut microbiota of Mediterranean loggerhead sea turtles have been pointed at as putatively capable of metabolizing pesticides (Arizza et al., 2019).
Research connecting microplastics ingestion with the gut microbiome is in its infancy (Fackelmann and Sommer, 2019). Studies mostly focused on model organisms and have been performed mostly in laboratory conditions, exposing model organisms at plastic concentrations and types that might not reflect the actual exposure. The mostly used model organisms incudes mouse (Lu et al., 2018; Jin et al., 2019) and zebrafish (Qiao et al., 2019; Gu et al., 2020; Kurchaba et al., 2020), or filter feeders, like mussels (Auguste et al., 2020; Li et al., 2020). In these studies, it was frequently reported that exposure to microplastics led to microbial communities distinct from the controls without microplastic treatments. This is likely due to the ability of plastic particles to be colonized by microbes on their surface (i.e., the so called “plastisphere”) (Amaral-Zettler et al., 2020), thus acting as potential carriers of microbial pathogens (Keswani et al., 2016). Moreover, PD can cause epithelial damages (Fackelmann and Sommer, 2019), that, in turn, may promote local inflammation and a possible response in the gut microbiome structure, that become more prone to colonization by opportunistic bacteria. Moreover, plastics can be vehicle of chemical pollutants, both adsorbed on their surface because of their hydrophobic nature and added to the plastic material itself during manufacturing (Galgani et al., 2014; Campanale et al., 2020), which can act as stressors, forcing changes on the composition of the intestinal microbial ecosystem.
In this scenario, the fecal microbiome emerges as a potential bioindicator for the impact of PD contamination on the physiology and health of living individuals of important marine flagship species, such as sea turtles (Foti et al., 2009; MSFD Technical Subgroup on Marine Litter Group et al., 2013; Galgani et al., 2014; Matiddi et al., 2017), allowing for an effective assessment of overall animal health.
To shed light on this perspective, we explored the amount of plastic debris found in the feces of live loggerhead sea turtles (Caretta caretta) rescued from the Northwestern Adriatic Sea, along with possible relationships between occurrence of PD and changes in the animal gut microbiota. Fecal samples were collected from sea turtles after their arrival at a rescue center, where veterinary attention and rehabilitation are provided to stranded, drifted or accidentally captured animals.
Loggerhead sea turtle is widely distributed in coastal tropical and subtropical waters around the world and quite common in the Mediterranean Sea (Márquez, 1990). Due to the great availability of food and warm shallow waters, the North Adriatic Sea is an important foraging and over-wintering area for Mediterranean loggerhead turtles (Franzellitti et al., 2004; Lucchetti and Sala, 2010). Although loggerheads in this area appear primarily threatened by the high rate of incidental bycatches during fishing activity (Lucchetti et al., 2017), pollution may represent an additional potential threat, as suggested by several evidence reporting well detectable tissue levels of widespread contaminants (Bucchia et al., 2015; Cocci et al., 2018, 2019, 2020). Here, the average benthic litter density has been estimated in 913 ± 80 items/km2 (Pasquini et al., 2016), ranking the Adriatic Sea as one of the most affected basin by plastic pollution worldwide. The selection of this site provides an exceptionally interesting model for studying the effect of plastic pollution on important flagship species such as sea turtles, considered as a holobiont, i.e., the animal and the microbes that live in a symbiotic relationship, in its whole complexity.
Materials and Methods
Study Animals and Samples Collection
The present study was conducted in the Northwestern Adriatic area, semi-enclosed shallow basin linked to the Mediterranean Sea, characterized by low salinity and strongly influenced by the Po river inputs. High temperature variations throughout the year are typical in this basin with an average depth of 35 m. The loggerhead sea turtles rescued in this area and involved in the present study, were found, stranded or captured by fishery nets, during winter and spring months from 2017 to 2019. Turtles were then temporarily hosted at the Sea Turtles Rescue Center “Ospedale delle Tartarughe–Fondazione Cetacea,” Riccione, Italy (43.99444°N; 12.673889°E). Turtles were kept in the center for cure and rehabilitation, hosted in single fiberglass tanks or tanks separated by a septum, fed twice a week with fishery products (small fishes such as pilchards, anchovies, or herrings, crustaceans, and mollusks provided by local fishermen active in the same Northwestern Adriatic area in which the turtles dwelled before being rescued), until release. The amount of fishes administered to each turtle is calculated by the veterinary personnel according to the animal size (0.11–0.12 kcal per g of individual weight for turtles with CCL < 30 cm, 0.04–0.05 kcal per g of weight for larger turtles).
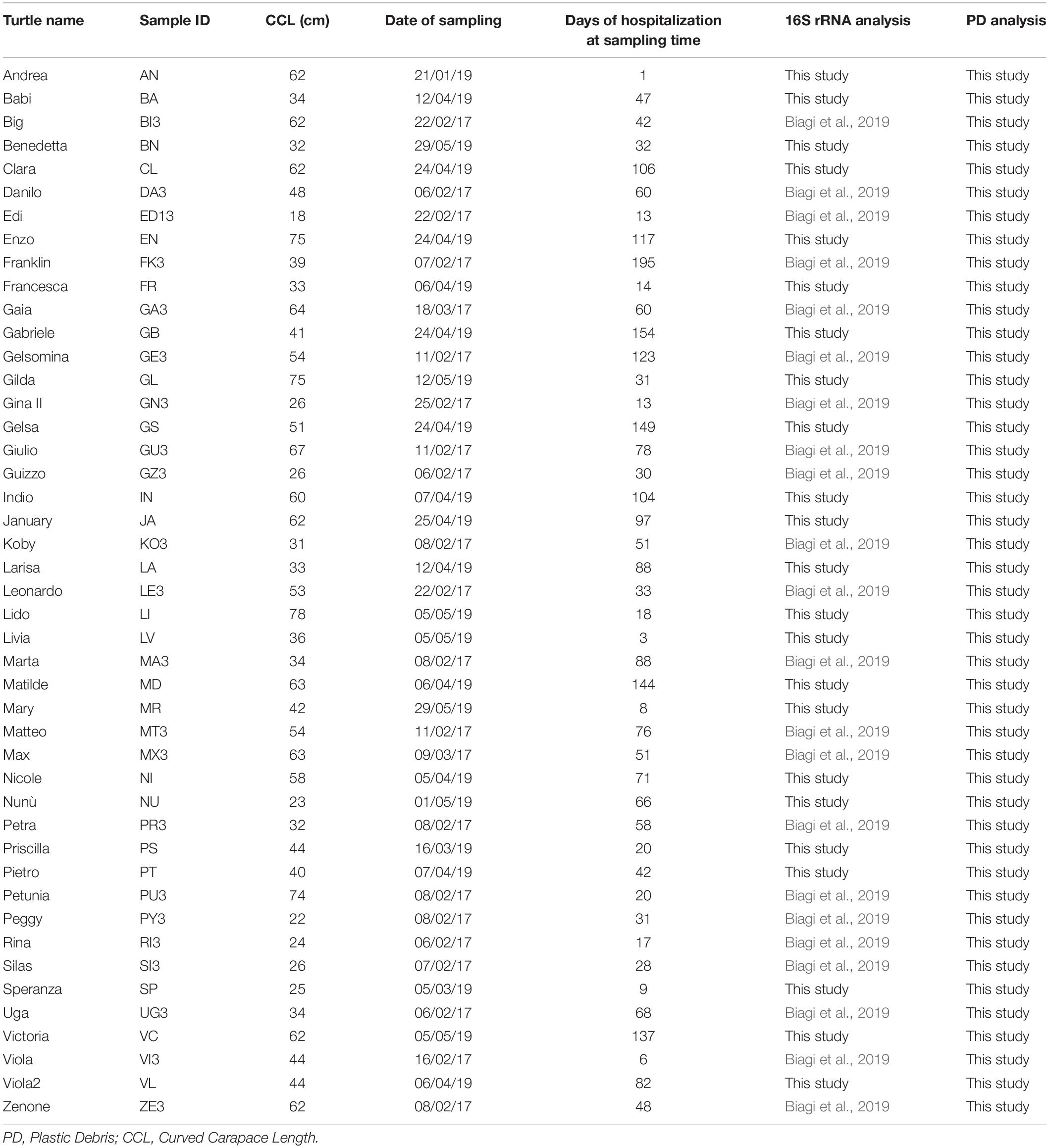
A total of 45 sea turtles were sampled for the present study. Turtle name, size (Curved Carapace Length, CCL), days of hospitalization at the sampling date, as well as samples ID and references are reported in Table 1. CCL was employed to determine life stage category of each individual, each corresponding to a preference for habitats (Casale et al., 2008, 2009, 2011): (i) size class = 28 cm (CCL range 0–28 cm): juveniles in the oceanic life stage; size class = 40 cm (CCL range 29–40 cm): juveniles undergoing through the transition from oceanic to neritic life; size class = 60 cm (CCL range 41–60 cm): sub-adults in oceanic and neritic stage; size class = 80 cm (CCL range 61–80 cm): sexually mature adults. CCL of assessed turtles ranged from 18 to 78 cm, and size class distribution is reported in Supplementary Table S1.
The first feces produced by each turtle after their arrival were collected from the tanks, using a metallic net that was washed each time twice using ultrapure Milli-Q water, and placed into sterilized glass containers. Feces were collected as soon as possible after production, trying to avoid prolonged contact with water, especially for what concern animals hosted in connected tanks. Volunteers working at the Rescue Center were trained on the importance to perform sampling correctly. Sampling was performed over a period of 5 months (January–May) in 2017 and in 2019. Because of the environmental and physiological stress that the enrolled turtles were enduring, they usually refused to eat for a variable length of time (as reported by Biagi et al., 2019). As a consequence, the first feces production happened over a very wide range of time after arrival, sometimes even weeks or months after hospitalization; this is highlighted in Table 1 where days of hospitalization at the time of fecal sampling is reported for each turtle. Samples were immediately frozen at −20°C, then transported to the laboratory using coolers with ice packs; samples were then stored at −80°C until further analysis, i.e., microbiota characterization and PD analysis. As highlighted in Table 1, for the 22 turtles sampled in 2017, 16S rRNA sequencing data were already available (Biagi et al., 2019, MG-Rast, https://www.mg-rast.org/linkin.cgi?project==mgp84794).
PD Extraction and Identification
After a literature research on the plastic debris recovery from animal fecal samples (Reynolds and Ryan, 2018; Hudak and Sette, 2019; Le Guen et al., 2020) in order to set up the best protocol, plastic particle extraction from turtles fecal samples was performed following the procedure reported by Valente et al. (2019). Briefly, a subsample of 0.2 g fecal material was weighed and placed in a glass beaker with 40 mL of 10% KOH for the degradation of the organic matter. The beaker was covered with a glass cap. Samples were incubated overnight at 40°C under continuous stirring. To minimize contaminations, the different steps of the protocol were carried out under a fume hood and all the surfaces were wiped down with ethanol, cotton lab coat and gloves were worn. All equipment used during the experiments were of glass rinsed with 10% HCl solution. Despite these precautions, contaminations could not be excluded. For this reason, a blank control sample containing only the extraction solution (40 mL of 10% KOH) was run in parallel to every set of analysis. Furthermore, filaments supposed to be of textile origin have been excluded from the analysis. Samples were pre-filtered using 1 mm sieves, and then filtered under vacuum filtration system through Whatman® glass microfiber filters, with 1.2 μm pore size. During the filtration, the filtration system was sealed with a glass dish to avoid contamination. Filters were placed in closed double glass dishes and covered with aluminum to protect them from light, because light exposure can lead to fragmentation of the polymers (Rios et al., 2007). Each filter was left to dry for 24 h at room temperature under the fume hood and inspected using a Nikon Eclipse 80i digital microscope. The entire surface was photographed with 4, 10, and 20X magnification with a digital camera (Digital Sight DS-2Mv, Nikon). Pictures were subsequently employed for particle counting and their classification according to shape and color. Particle size (maximum linear dimension for filaments or irregular shapes; maximum diameter for rounded and angular shapes) was assessed using the ImageJ image analysis software. Negative controls were compared with the samples, thus allowing to detect possible contaminations within each set of analysis.
Microbial DNA Extraction
Total DNA extraction from fecal samples was carried out using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) with a modified protocol as previously reported by Biagi et al. (2019). DNA was then quantified by using NanoDrop ND1000 (NanoDrop Technologies, Wilmington, DE) and stored at −20°C for subsequent processing. Extracted DNA was diluted to the final concentration of 5 ng/μl using PCR grade water, immediately before performing PCR amplification. The V3–V4 hypervariable region of the 16S rRNA gene was PCR-amplified using the 341F and 785R primers with added Illumina adapter overhang sequences, as previously described (Barone et al., 2019). Five microliter of diluted DNA were used as template for PCR in a final volume of 50 μl. PCR reactions were purified by using Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA). Indexed libraries were then prepared by limited-cycle PCR reaction, using Nextera technology (Illumina, San Diego, CA). Libraries were normalized to 4 nM and pooled, after a further clean-up step as described above. The sample pool was finally denatured with 0.2 N NaOH and diluted to 6 pM with a 20% PhiX control. Sequencing was performed on Illumina MiSeq platform using a 2 × 250 bp paired end protocol, according to the manufacturer’s instructions (Illumina, San Diego, CA). Sequencing reads were deposited in SRA-NCBI (project number PRJNA679693).
Bioinformatics and Statistics
For PD occurrence and distribution among animals, the datasets were analyzed permutation multivariate analysis of variance (PERMANOVA) using PRIMER v6 (Anderson et al., 2008). Log- transformed data were used to calculate similarity matrices based on the Euclidean distance (999 permutations; P(perm) < 0.05). Correlation analyses (Spearman’s test), data visualization, and graphics were obtained with the GraphPad Prism software ver 9. In any case, statistical differences were accepted when P < 0.05. PD size distribution among different animal size classes and different PD shapes was analyzed by non-parametric one-way ANOVA (Kruskal-Wallis test) followed by the Mann-Whitney U-test, after deviations from parametric ANOVA assumptions being assessed (Normality: Shapiro-Wilk’s test; equal variance: F-test). These statistical analyses were performed using the GraphPad Prism 9 software (GraphPad Inc.).
Raw sequences were processed using a pipeline combining PANDAseq (Masella et al., 2012) and QIIME (Caporaso et al., 2010). High-quality reads were binned into operational taxonomic units (OTUs) according to the taxonomic threshold of 97% using UCLUST (Edgar, 2010). The 97%-similarity threshold allowed us to obtain groups of sequences, possibly ascribable to species or small group of species, that could play specific ecological roles in the ecosystem, as previously reported (Biagi et al., 2020). The taxonomy was assigned using the RDP classifier against the Greengenes database (release May 2013). Unassigned sequences and those assigned to chloroplasts and mitochondria were discarded. The PD amount found in each fecal sample was normalized to number of debris per 10 g of stool (Schwabl et al., 2019). Samples were then split into 3 groups according to the normalized PD content: PD < 100, PD 101–300, and PD > 300 counts/10 g of stool. Samples included in each group and average features of the corresponding animals are reported in Table 2. Statistical analyses were performed using R version 3.5.11 and the packages stats, gplots, vegan, and made4. The similarity percentage (SIMPER) analysis (Clarke, 1993) was carried on to determine the contribution from individual OTUs to the dissimilarity between the three groups of samples (PD < 100, PD 101–300, PD > 300), using the function simper in the vegan package of R. OTUs with a p-value < 0.05 in at least two of the contrasts (PD < 100 vs. PD 101–300, PD < 100 vs. PD > 300, PD 101–300 vs. PD > 300) were retained for the subsequent clustering analysis in order to focus on the microbiome features possibly responding to a PD dose effect. Representative sequences of the OTUs of interest were identified as the highest score alignment to the NCBI bacterial 16S rRNA gene database (release September 2019) using BLASTn (version 2.9.0). Prevalence of each OTU in the groups of samples was calculated as percentage of samples in each group in which the reads assigned to the selected OTU were detected (relative abundance > 0). Correlation between OTUs relative abundance and continuous variables (i.e., normalized PD amount, CCL values) was calculated as Kendall correlation tau values; significance of the correlation was calculated using the function cor.test in the package stats in R. Sample clustering was performed accordingly to the selected OTUs relative abundance, adopting Spearman correlation coefficients as metric and Ward-linkage method. Heatmap was produced using the function heatmap.2 in R.
Results
PD Occurrence, Concentration, and Size Distribution
All individuals except one (MT; Figure 1) showed particle items in the feces, for an average value of 6 ± 6.09 particles/sample of feces (0.2 g) (Figure 1). The maximum number of particles found in a single individual is 34. Frequency distribution of the different classes of particle occurrence shows that the most frequent number of items per sample was 1 (n = 15) and 2 (n = 14) (Insert in Figure 1). No significant relationship was observed between particle sample content and hospitalization time, i.e., the time during the hospitalization at which the sample was collected (r = −0.32, P > 0.05; Spearman correlation test).
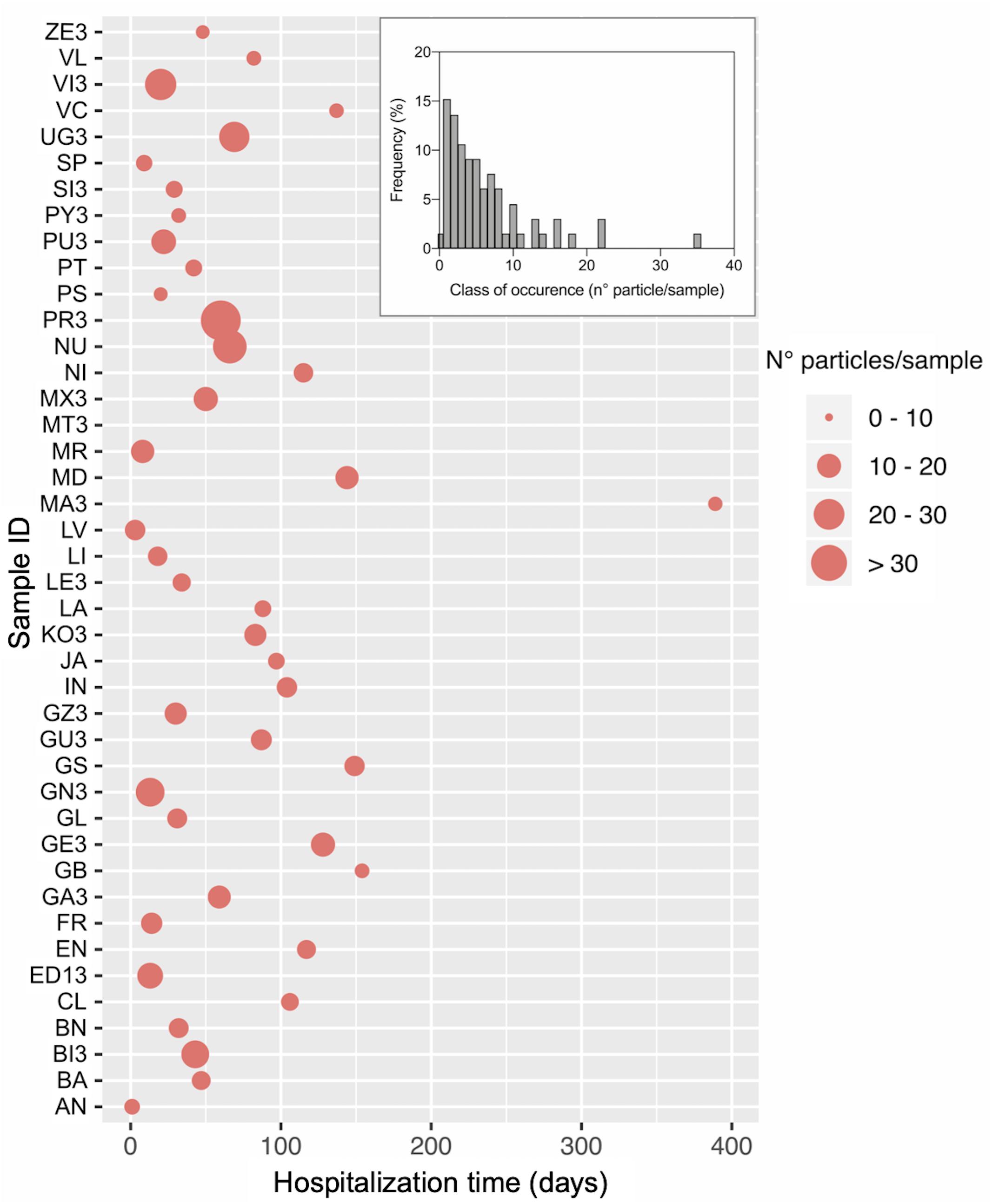
Figure 1. Number of plastic particles isolated from fecal samples of loggerhead sea turtles rescued in the Northwestern Adriatic Sea. The main graph shows the number of particles for each in individual and the day within the hospitalization period at which the sample was collected. The Insert reports the frequency distribution of the number of isolated particles. Sample ID and total hospitalization period are as reported in Table 1.
Particle classification according to color and shape is reported in Figure 2. In all size class categories, data show the prevalence of filaments, followed by unclassified shapes, angular fragments, and finally round items (Figure 2H). As to colors, the most represented classes are transparent/white and red particles (Figure 2G). There were no statistical differences for shape or color category frequencies amongst different size classes (P(perm) > 0.05; Supplementary Table S2). PD size ranged between 11 and 889 μm with a mean value of 198 μm. No macrodebris (PD > 1 mm) was detected in our samples, likely due to the animal digestive processes. PD size distribution between turtle size classes was not statistically significant (Figure 2J upper panel), whereas within different particle shape category (Figure 2J, lower panel), filaments resulted significantly longer than angular or other shape PD [(mean, min-max range): filaments = 304 μm, 63–889 μm; angular = 59.6 μm, 12–153 μm; others = 86.4 μm, 11–212 μm; P < 0.05].
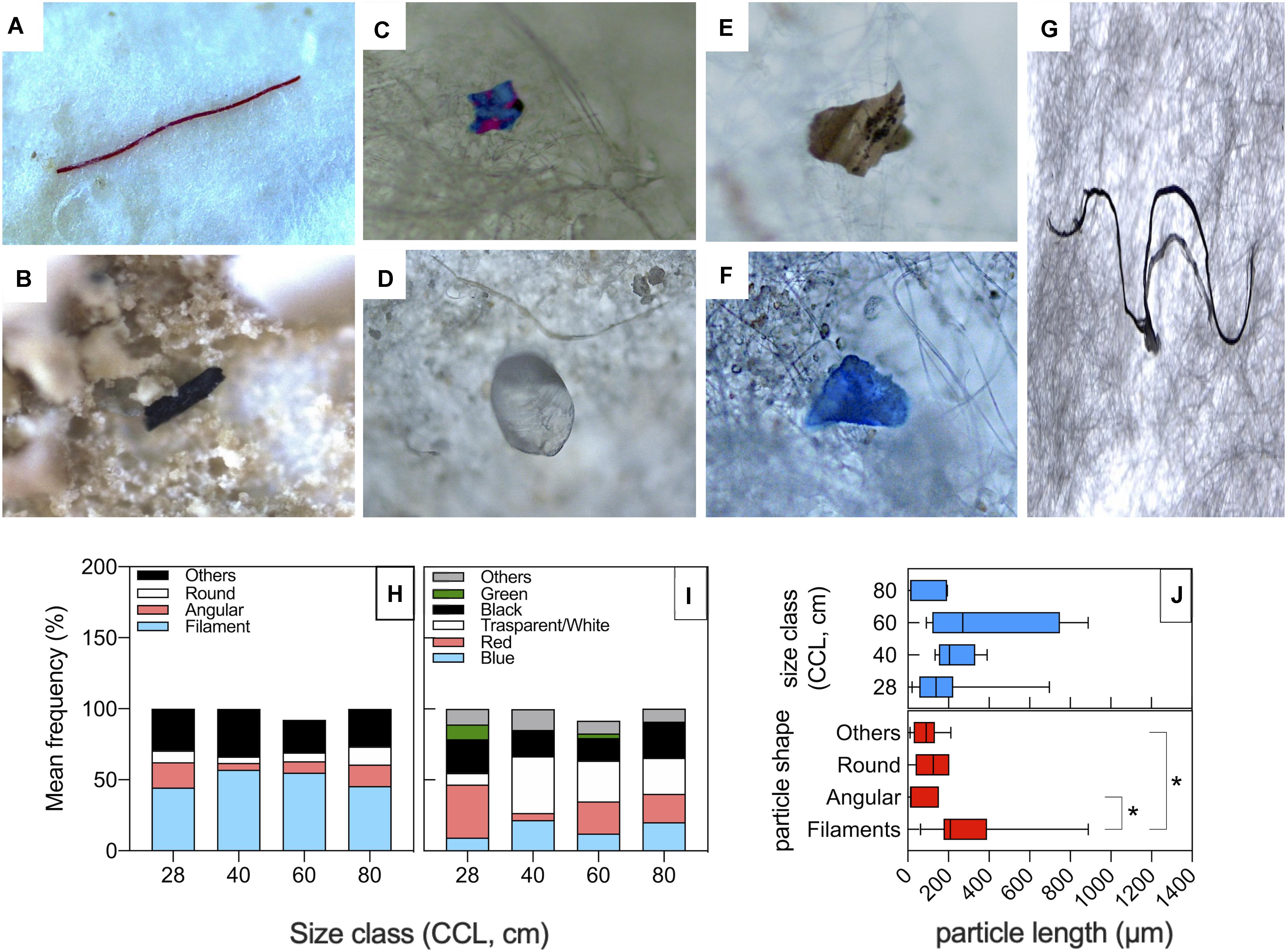
Figure 2. Shapes and colors of particles isolated from fecal samples of loggerhead sea turtles from the Northwestern Adriatic Sea. (A–G) Microscope pictures showing representative particle items isolated from the sea turtle fecal samples: (A) Red filaments; (B) Black angular fragments; (C) Angular fragments with unclassified color (others); (D) Transparent round fragments; (E) Unclassified shape and color; (F) Blue fragment with unclassified shape; (G) Black filaments. Average frequency classification by shape (H) and by color (I) across different turtle size classes. (J) PD size distribution among turtle size classes (upper panel) and particle shape category (lower panel). *P < 0.05.
Impact of PD on the Sea Turtle Gut Microbiota
The fecal microbiota structure of the 23 sea turtles sampled in 2019 was profiled by NGS of the V3–V4 hypervariable region of the 16S rRNA gene. Reads from the present study were analyzed together with those obtained from the 22 sea turtles available from Biagi et al. (2019). A total of 1,884,864 paired-end sequences passed quality filtering (mean per sample ± SD, 41 886 ± 17 972). High-quality reads were clustered into 19 252 operational taxonomic units (OTUs).
To explore peculiarities of microbiota composition in sea turtles related to the PD content, samples were grouped according to PD amounts normalized to 10 g of stool, as previously reported (Schwabl et al., 2019): group “PD < 100” included 16 samples with a PD content < 100 counts per 10 g of stool, group “PD 101–300” included 13 samples with a PD content ranging from 101 to 300 counts per 10 g of stool, and group “PD > 300,” with the remaining 16 samples counting > 300 PD per 10 g of stool (Table 2). The distribution of turtles’ size was homogeneous among PD content groups (Kruskal-Wallis test, P > 0.05). Also, groups based on PD content did not differ significantly in terms of days of hospitalization at the time of sampling (P > 0.05).
Similarity percentage (SIMPER) analysis was carried out to identify the best subset of OTUs contributing to the dissimilarity in microbiota between groups of samples with different PD content. In order to focus our analysis on those microbiome features whose abundance increased or decreased in association to the increasing concentration of PD in the feces, we selected OTUs significantly (simper analysis P-value < 0.05) contributing to the dissimilarity between groups in at least two of the contrasts (PD < 100 vs. PD 101–300, PD < 100 vs. PD > 300, PD 101–300 vs. PD > 300). Fifty-eight OTUs emerged from this selection, 48 with an increasing abundance and/or prevalence in association with the normalized PD amount, and 9 showing an opposite trend (Table 3). The cumulative contribution of the selected OTUs to the dissimilarity between groups was 0.6% for PD < 100 vs. PD 101–300 contrast, 1.6% for PD < 100 vs. PD > 300 contrast, and 1.5% for PD 101–300 vs. PD > 300, showing that the PD content accounted for a minor quota of the microbiome dissimilarity among the animals. To confirm the association of the identified OTUs to the PD content, Kendall correlation coefficient (tau) was calculated between the relative abundance of each OTUs and the normalized PD content of the corresponding samples; correlation tendencies (tau > 0.40 or < −0.40) with P-values < 0.01 are shown in Table 3. Since PD content may be affected by feeding preferences of sea turtles, that largely depend on life stage (i.e., size class category as reported in Supplementary Table S1), we sought for a possible relationship between PD content, size and OTU distribution. Kendall tau was also calculated using CCL measurements as reference variable, as control of specificity for the selected OTUs as PD content responders. None of the selected OTUs was significantly correlated to the CCL of the corresponding animal (Supplementary Table S3).
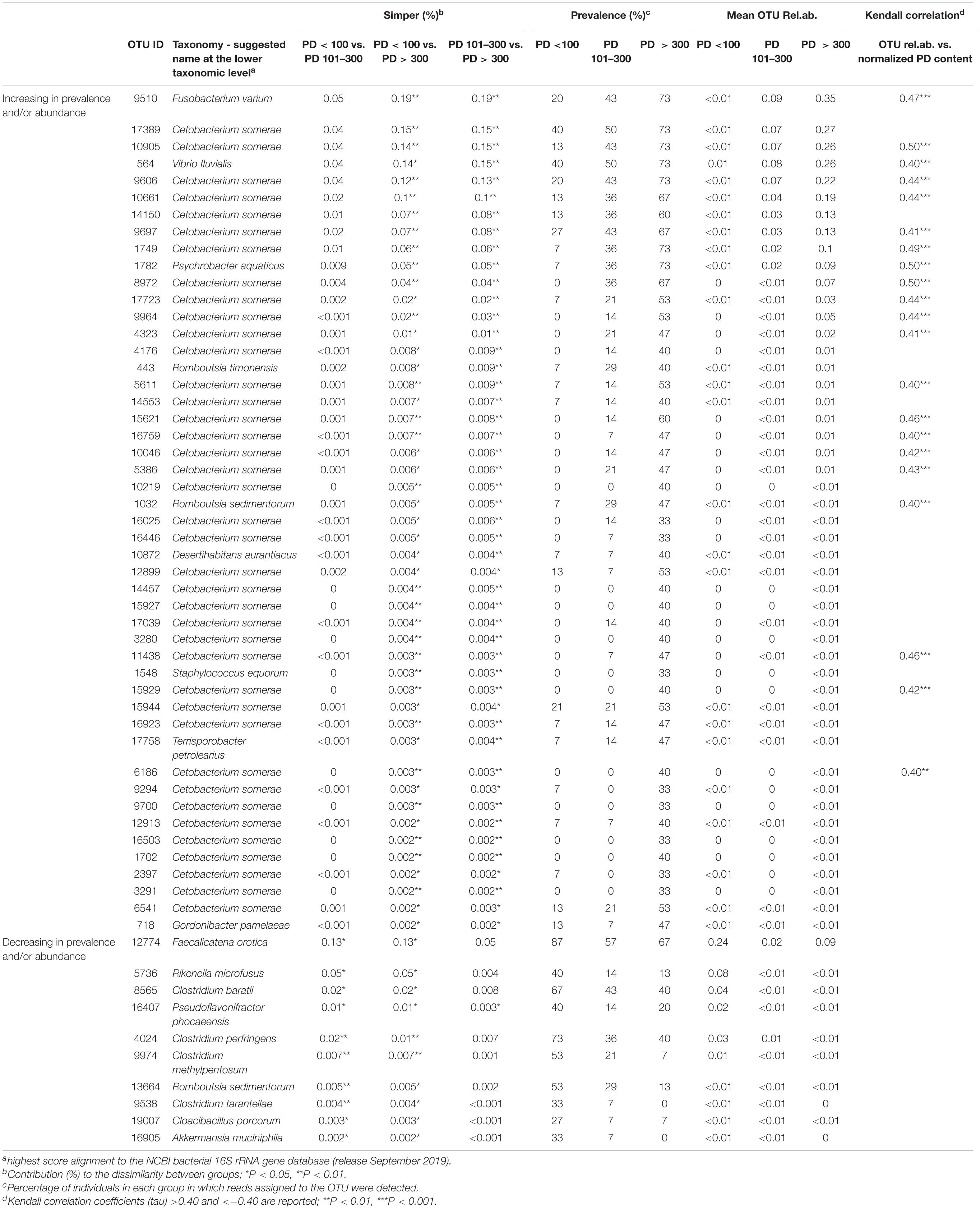
Table 3. Taxonomy, abundance, and prevalence of OTUs significantly contributing to the dissimilarity among PD < 100, PD101–300, and PD > 300 groups of samples.
The 58 OTUs, mostly contributing to the dissimilarity among PD content groups were identified down to species level as the highest score alignment to the NCBI bacterial 16S rRNA gene database (Table 3). Thirty-nine out of 48 OTUs showing an increasing trend in association to the PD content were assigned to the species Cetobacterium somerae. However, among the most contributing OTUs we found also two sequences assigned to animal pathogens, namely Fusobacterium varium (Duangurai et al., 2019) and Vibrio fluvialis (Arab et al., 2020). Other OTUs in this subset were assigned to the following genera: Psychrobacter, Romboutsia, Desertihabitans, Staphylococcus, Terrisporobacter, and Gordonibacter.
The 9 OTUs that showed a decreasing trend associated with the increasing PD content in feces were assigned to different microorganisms, several of which belonging to the genus Clostridium, with representation of different species. The one contributing most (0.13%) to the dissimilarity between PD < 100 samples and the other two groups was assigned to the species Faecalicatena orotica, an animal gut isolate previously classified as Clostridium oroticum (Sakamoto et al., 2017). Other genera represented in this group of OTUs were Akkermansia, Rikenella, Cloacibacillus, Pseudoflavonifractor, and Romboutsia, all previously identified in vertebrate gut bacterial ecosystem.
To confirm that the identified OTUs were discriminant for the content of PD in corresponding fecal samples, we performed a cluster analysis based on Spearman correlation between the abundance profiles of these selected 58 OTUs (Figure 3). Even if a certain level of dispersions was maintained, samples showed an overall tendency toward the segregation between groups, defining two major clusters, with samples from the PD > 300 group preferentially assigned to the cluster A, particularly enriched in those OTUs identified by the simper analysis as significantly contributing to the dissimilarity between PD > 300 samples and both the other groups. On the contrary, PD < 100 samples grouped together in cluster B, characterized by an overall higher abundance of the OTUs identified by the simper analysis as significantly contributing to the dissimilarity between PD < 100 samples and both the other groups (lower part of the heatmap). Samples from the intermediate group (PD 101–300) were distributed equally across both clusters.
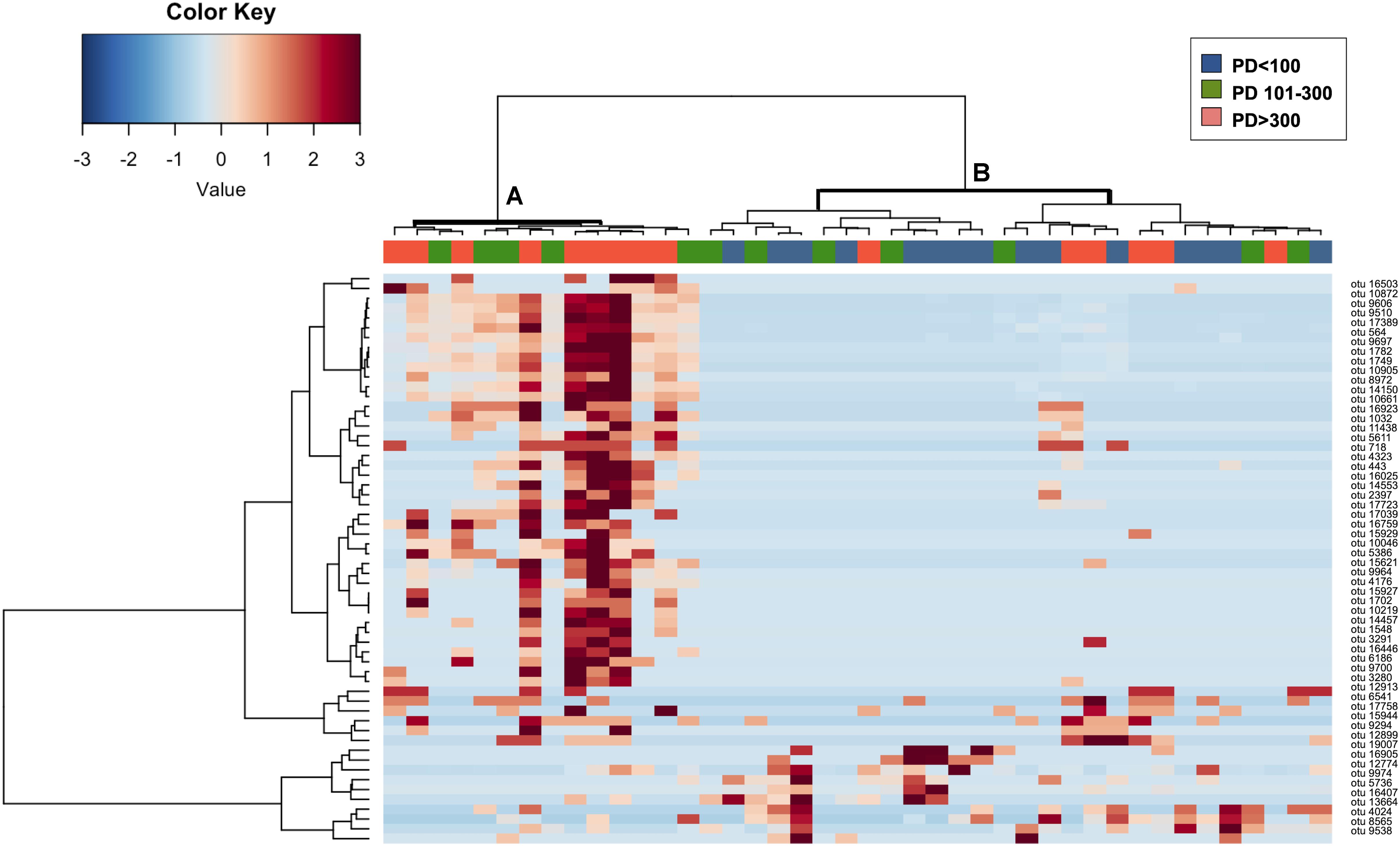
Figure 3. Hierarchical clustering and relative abundance of OTUs significantly contributing to the separation of samples in PD < 100, PD 101–300 and PD > 300 groups. The heatmap shows Ward-linkage clustering based on the Spearman correlation coefficients of the available sea turtles fecal samples and 58 fecal OTUs that were found significantly contributing to the separation of samples into the three PD content groups according to the simper analysis. Samples are shown column-wise; color code (blue, green, and orange) is provided for identifying samples belonging to groups PD < 100, PD101–300, and PD > 300. OTUs relative abundances are reported on the rows. OTUs are identified as numbers; taxonomical assignation based on the highest score alignment to the NCBI 16S rRNA gene database is provided in Table 3. The two identified samples clusters (clusters A and B) are highlighted in bold.
Discussion
The present study explored the plastic debris (PD) amount in the feces of wild captured loggerhead sea turtles (C. caretta), dwelling in the Northwestern Adriatic Sea, collected after their arrival at a local rescue center for their rehabilitation. Large marine vertebrates, such as cetaceans and sea turtles, are ideal sentinels to monitor plastic pollution in marine environment but, thus far, data on the impact of plastic ingestion on such wild species have been necessarily provided mostly through sampling of gastrointestinal contents from dead animals (López-Martínez et al., 2020). By employing fecal samples, our approach attempts to assess the actual distribution of PD in the gastrointestinal tract of live animals at different life stages, although it is challenging to compare our results with the available literature. The number of plastic particles found in the feces of the turtles involved in our study can be considered as quite high, with respect to data generally reported for the gastrointestinal content of dead stranded turtles. For instance, Duncan et al. (2018) found an average plastic micro-particles content ranging between 10 and 15 per 100 ml of gastric content of both C. caretta and Chelonia mydas from the Mediterranean Sea. Such number, even if higher than those reported for the same species of turtles recovered from Atlantic and Pacific oceans in the same paper, is still very low in comparison to the PD counts reported in our work, normalized to 10 g of solid feces. The comparison between the present study and previous ones is challenging because of the different nature of the matrix (liquid gastrointestinal contents vs. solid feces) and the different counting approach (normalization to 10 g of feces vs. counting of plastic items per samples or percentage of samples contaminated by plastic debris (Camedda et al., 2014; Hoarau et al., 2014). However, it is tempting to hypothesize that the peculiar location of our study, the Northwestern Adriatic Sea, might be partially responsible of the discrepancy between results, and for the fact that all, but one, of the 45 enrolled turtles provided fecal samples containing PD. The Adriatic basin is, in fact, one of the most polluted marine sites across the globe, due to its high productivity and anthropic impact, with an average concentration of > 400,000 plastic particles up to 5 mm per km2 (MSFD Technical Subgroup on Marine Litter Group et al., 2013; Alessi and Di Carlo, 2018; Llorca et al., 2020). Renzi et al. (2018) assessed the presence of PD in Mytilus galloprovincialis bred in a mussel farm off Cesenatico (Northwestern Adriatic Sea, Italy), in comparison with mussels bred in other Italian coastal areas and with natural populations of the Tyrrhenian Sea. The concentration of PD in animals from the Northwestern Adriatic resulted higher than that of other areas, including individuals residing in nature, with an average of 12.4 items/mussel. All the materials found in the sampled individuals were filaments, with a dominance of blue and black colors. Bivalve mollusks are one of the main components of the loggerheads diet, and it was demonstrated that they can transfer particles to other invertebrates such as crabs (Farrell and Nelson, 2013), which are also a prey of C. caretta. Therefore, showing a relevant level of plastic pollution in fecal material of Adriatic Sea turtles—and in the light of the recognized relevance of sea turtle as flag species for the health status of the marine environment—these results indicate and confirm the high level of plastic pollution in Adriatic ecosystems.
Besides representing a flagship species for the levels of environmental contamination, sea turtle health may be directly impacted by plastic ingestion. Larger debris have macroscopic effects, for instance by inducing gastric blockage and injuries, but also small fragments are known to affect turtle physiology (Bugoni et al., 2001; Derraik, 2002; Kühn et al., 2015; Nicolau et al., 2016). The impact of PD on the turtle health can also be mediated by the interaction with the intestinal microbial community, the composition of which can be altered by plastic-associated chemicals (i.e., adsorbed pollutants or plastic additives), and/or biological contaminants (i.e., plastisphere microbial components) (Amaral-Zettler et al., 2020; Campanale et al., 2020). Probably because of the difficulty in obtaining samples from wild animals, studies connecting microbiome composition and plastic gastrointestinal contamination have been mostly limited to model animals or easily accessible productive species, such as bivalves of interests for food consumption (Lu et al., 2018; Jin et al., 2019; Qiao et al., 2019; Auguste et al., 2020; Gu et al., 2020; Kurchaba et al., 2020; Li et al., 2020). Interestingly, in our observational study on wild capture animals only one of the turtles provided a sample free of fecal PD, mirroring the pervasive real world plastic contamination in the particularly polluted geographical location. Consequently, we choose to focus our attention on the effect of the PD amount on the fecal microbiota composition. Indeed, our statistical approach based on the analysis of similarity percentages (SIMPER) identified several microbial sequences whose abundance significantly contributed to the dissimilarity between groups of samples with different PD content. The cumulative contribution of the identified OTUs to the dissimilarity between groups of samples was expectedly small, since other variables (e.g., diet, health, age, individual physiology, history of environmental exposition) are known to play a relevant role in defining the overall structure the microbiome in animals. Still, we were able to define two groups of PD-responding microbiome OTUs that contributed to effectively separate the samples with the highest PD content from those with the lower one. We defined a first group of 48 OTUs, mainly composed by sequences assigned to the species Cetobacterium somerae, which showed an increasing trend, in terms of relative abundance and/or prevalence in the sample groups, along with the increasing PD content in feces. Conversely, a second, more diverse OTUs list was obtained showing an opposite trend.
C. somerae belongs to the Fusobacteria phylum and has already been reported as abundant in the gut microbiota of Adriatic loggerhead sea turtle (Biagi et al., 2019). In particular, this species is known to ferment amino acids and peptides into short-chain fatty acids (SCFAs) that can be absorbed and utilized by both bacteria and the host (Tsuchiya et al., 2008; Olsen, 2014) and it also benefits the host by producing vitamin B12 and antimicrobial peptides (Ahasan et al., 2018). The reason why the highest PD content was associated to higher abundance of 16S reads assigned to this species is to be explored further in future studies. Recent studies on fish models (marine manaka and crucian carp) showed that Cetobacterium amount responds peculiarly to increasing exposure to organic chemical pollutants (perfluorobutanesulfonate and ammonia, respectively): exposure to low concentration of pollutants caused an increase in Cetobacterium, while its abundance dropped dramatically after exposure to high concentration (Qi et al., 2017; Chen et al., 2018). Perfluorobutansulfonate (PFBS) is used as processing aids in the manufacture of fluoropolymers and as flame retardants in polycarbonate materials (ECHA, 2019). It is not unthinkable that, even if it is usually considered a symbiont for marine carnivorous species, the increase in bacteria assigned to the genus Cetobacterium in our study could be actually related to some contaminants added in low concentration on the plastic materials. Moreover, microplastic contamination has been associated to an increase ammonia excretion in a model fish (Yin et al., 2019). Alongside the majority of Cetobacterium-assigned sequences, the OTUs showing an increasing pattern associated with the increasing PD content in feces also included some sequences assigned to bacterial species previously isolated from petroleum-polluted environmental sites, i.e., Terrisporobacter petrolearius (Deng et al., 2015), as well as potentially pathogenic species (i.e., Vibrio fluvialis and Fusobacterium varium) (Duangurai et al., 2019; Arab et al., 2020). The latter observations can support the hypothesis that PD can act as carriers for pollutant-selected and potential pathogenic species, easing their entrance and colonization of the gastrointestinal environment of marine animals (Fackelmann and Sommer, 2019).
The OTUs showing higher abundance or prevalence in the group of samples with lower PD content constituted a more diverse collection, all assigned to genera that were isolated from and/or are commonly found in vertebrate gastrointestinal tract, such as Akkermansia, Rikenella, Faecalicatena, Cloacibacillus, and Clostridium (Costello et al., 2010; Looft et al., 2013; Sakamoto et al., 2017; Biagi et al., 2019; Lin et al., 2019). It might be postulated that an increased amount of PD accumulating in the lower gastrointestinal tract might result in a less favorable environment for the survival of some endogenous symbiotic species, possibly because of the augmented local inflammation caused by small epithelial injuries or through effects of plastic-associated contaminants (Fackelmann and Sommer, 2019).
In conclusion, our observational study represents a step forward into the research field of evaluating the impact of plastic contamination in the marine environment. Indeed, beside involving ecologically relevant, wild, alive animals that did not undergo controlled plastic contamination, as reported in very few previous studies (Camedda et al., 2014; Hoarau et al., 2014), our work has the unprecedented feature of contextually exploring both PD content and microbiome composition of sea turtles fecal samples. Even if a partial influence of the controlled environment in which turtles were temporarily kept cannot be completely ruled out, the level of plastic contamination of the turtles is likely to reflect that of the marine food web of the Adriatic Sea. Indeed, the amount of PD found in the turtle feces was not statistically correlated to the number of days the turtles spent in captivity before sampling, possibly confirming that plastics are highly persistent and accumulative in the digestive tract of these animals, an aspect that could not be explored in previous studies on dead stranded animals. To date, standard methodologies and criteria of quantification have been provided to correlate the plastic litter found in dead turtles to the environmental status (Matiddi et al., 2019). The results provided by our analysis cannot be compared to such standard guidelines because of the differences in methodological approach. However, the detection and quantification of PD in feces of rescued, live animals, as presented in our work, highlighted that loggerheads’ fecal samples might become a good proxy to evaluate the level of threat constituted by the PD in the Adriatic Sea, to be used together with other standardized surveys. Future research involving sea turtles from rescue centers located in different, and differently polluted, geographic area could provide data to sustain the potential of this approach.
For what concern the relationship between fecal microbial composition and PD concentration in fecal material, the detected “dose effect” of fecal plastic contamination regarded, as foreseeable, minor components of the microbial community. Still, the data presented here identified some putative gut microbiome biomarkers of plastic contaminations, such as a selection of microbiome components whose abundance increases in association to plastic-associated chemical pollutants (plastic flame retardants or other plastic components) and the presence of potential environmental pathogens potentially enriched on the plastisphere. Further, PD contamination was also associated with the depletion in several putative mutualistic components of the gut microbiome. Our findings open the way to future studies exploring the impact of the observed gut microbiome changes in terms of mechanism of action and consequent effects on the animals’ health and physiology.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/ PRJNA679693.
Ethics Statement
Ethical review and approval was not required for the animal study because Loggerhead sea turtles feces were collected from animals temporarily hosted at a local, authorized Rescue Center for veterinary attention before being released in the sea. Fecal samples were collected from tank water without touching nor disturbing the animals in any way.
Author Contributions
SF and MC: conceptualization, writing—review and editing, and supervision. EB, SR, SF, and MC: methodology. EB, MM, GP, VA, CR, DS, and SF: investigation. EB, MM, SP, SR, and SF: formal analysis. EB, MM, CR, and SF: data curation. VA, SP, SF, and MC: resources. EB, MM, and SF: writing—original draft preparation. SP, SF, and MC: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No. 818290. This publication reflects only the author’s view and the Agency is not responsible for any use that may be made of the information it contains.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study represents partial fulfillment of the requirements for the Ph.D. thesis of MM, GP, and DS at the Ph.D. course of Innovative Technologies and Sustainable Use of Mediterranean Sea Fishery and Biological Resources (FishMed—University of Bologna, Italy).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.637030/full#supplementary-material
Supplementary Table S1 | Size classification and distribution of the loggerhead sea turtles assessed in this study.
Supplementary Table S2 | Results of PERMANOVA pairwise comparisons of particle shape/color distribution among different sea turtle size classes.
Supplementary Table S3 | Kendall correlation coefficient (tau) and associated P-values calculated between the relative abundance of each OTU (Table 1) significantly contributing to the samples grouping and the normalized PD content and CCL of the corresponding turtles.
Footnotes
References
Ahasan, M. S., Waltzek, T. B., Huerlimann, R., and Ariel, E. (2018). Comparative analysis of gut bacterial communities of green turtles (Chelonia mydas) pre-hospitalization and post-rehabilitation by high-throughput sequencing of bacterial 16S rRNA gene. Microbiol. Res. 207, 91–99. doi: 10.1016/j.micres.2017.11.010
Alessi, E., and Di Carlo, G. (2018). Out of the Plastic Trap: Saving the Mediterranean from Plastic Pollution. Rome: WWF Mediterranean Marine Initiative Report.
Amaral-Zettler, L. A., Zettler, E. R., and Mincer, T. J. (2020). Ecology of the plastisphere. Nat. Rev. Microbiol. 18, 139–151. doi: 10.1038/s41579-019-0308-0
Anderson, M., Gorley, R. N., and Clarke, K. (2008). PERMANOVA+ for Primer: Guide to Software and Statistical Methods. Plymouth: PRIMER-E, Ltd.
Arab, S., Nalbone, L., Giarratana, F., and Berbar, A. (2020). Occurrence of Vibrio spp. Along the algerian mediterranean coast in wild and farmed sparus aurata and dicentrarchus labrax. Vet. World. 13, 1199–1208. doi: 10.14202/vetworld.2020.1199-1208
Arizza, V., Vecchioni, L., Caracappa, S., Sciurba, G., Berlinghieri, F., Gentile, A., et al. (2019). New insights into the gut microbiome in loggerhead sea turtles Caretta caretta stranded on the Mediterranean coast. PLoS One 14:e0220329. doi: 10.1371/journal.pone.0220329
Auguste, M., Lasa, A., Balbi, T., Pallavicini, A., Vezzulli, L., and Canesi, L. (2020). Impact of nanoplastics on hemolymph immune parameters and microbiota composition in Mytilus galloprovincialis. Mar. Environ. Res. 159:105017. doi: 10.1016/j.marenvres.2020.105017
Barboza, L. G. A., Dick Vethaak, A., Lavorante, B. R. B. O., Lundebye, A. K., and Guilhermino, L. (2018). Marine microplastic debris: an emerging issue for food security, food safety and human health. Mar. Pollut. Bull. 133, 336–348. doi: 10.1016/j.marpolbul.2018.05.047
Barnes, D. K. A., Galgani, F., Thompson, R. C., and Barlaz, M. (2009). Accumulation and fragmentation of plastic debris in global environments. Philos. Trans. R. Soc. B Biol. Sci. 364, 1985–1998. doi: 10.1098/rstb.2008.0205
Barone, M., Turroni, S., Rampelli, S., Soverini, M., D’Amico, F., Biagi, E., et al. (2019). Gut microbiome response to a modern Paleolithic diet in a Western lifestyle context. PLoS One 14:e0220619. doi: 10.1371/journal.pone.0220619
Biagi, E., D’Amico, F., Soverini, M., Angelini, V., Barone, M., Turroni, S., et al. (2019). Faecal bacterial communities from Mediterranean loggerhead sea turtles (Caretta caretta). Environ. Microbiol. Rep. 11, 361–371. doi: 10.1111/1758-2229.12683
Biagi, E., Mengucci, C., Barone, M., Picone, G., Lucchi, A., Celi, P., et al. (2020). Effects of vitamin B2 supplementation in broilers microbiota and metabolome. Microorganisms 8:1134. doi: 10.3390/microorganisms8081134
Bjorndal, K. A., Bolten, A. B., and Lagueux, C. J. (1994). Ingestion of marine debris by juvenile sea turtles in coastal Florida habitats. Mar. Pollut. Bull. 28, 154–158. doi: 10.1016/0025-326X(94)90391-3
Bucchia, M., Camacho, M., Santos, M. R., Boada, L. D., Roncada, P., Mateo, R., et al. (2015). Plasma levels of pollutants are much higher in loggerhead turtle populations from the Adriatic Sea than in those from open waters (Eastern Atlantic Ocean). Sci. Total Environ. 523, 161–169. doi: 10.1016/j.scitotenv.2015.03.047
Bugoni, L., Krause, L., and Petry, M. V. (2001). Marine debris and human impacts on sea turtles in Southern Brazil. Mar. Pollut. Bull. 42, 1330–1334. doi: 10.1016/S0025-326X(01)00147-3
Camedda, A., Marra, S., Matiddi, M., Massaro, G., Coppa, S., Perilli, A., et al. (2014). Interaction between loggerhead sea turtles (Caretta caretta) and marine litter in Sardinia (Western Mediterranean Sea). Mar. Environ. Res. 100, 25–32. doi: 10.1016/j.marenvres.2013.12.004
Campanale, C., Dierkes, G., Massarelli, C., Bagnuolo, G., and Uricchio, V. F. (2020). A relevant screening of organic contaminants present on freshwater and pre-production microplastics. Toxics 8:E100. doi: 10.3390/toxics8040100
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–336. doi: 10.1038/nmeth.f.303
Caron, A. G. M., Thomas, C. R., Berry, K. L. E., Motti, C. A., Ariel, E., and Brodie, J. E. (2018). Ingestion of microplastic debris by green sea turtles (Chelonia mydas) in the great barrier reef: validation of a sequential extraction protocol. Mar. Pollut. Bull. 127, 743–751. doi: 10.1016/j.marpolbul.2017.12.062
Casale, P., Abbate, G., Freggi, D., Conte, N., Oliverio, M., and Argano, R. (2008). Foraging ecology of loggerhead sea turtles Caretta caretta in the central Mediterranean Sea: evidence for a relaxed life history model. Mar. Ecol. Prog. Ser. 372, 265–276. doi: 10.3354/meps07702
Casale, P., d’Astore, P. P., and Argano, R. (2009). Age at size and growth rates of early juvenile loggerhead sea turtles (Caretta caretta) in the Mediterranean based on length frequency analysis. Herpetol. J. 19, 29–33.
Casale, P., Mazaris, A. D., and Freggi, D. (2011). Estimation of age at maturity of loggerhead sea turtles Caretta caretta in the Mediterranean using length-frequency data. Endangered. Species Res. 13, 123–129. doi: 10.3354/esr00319
Chen, L., Lam, J. C. W., Hu, C., Tsui, M. M. P., Wang, Q., Giesy, J. P., et al. (2018). Perfluorobutanesulfonate exposure causes durable and transgenerational dysbiosis of gut microbiota in marine medaka. Environ. Sci. Technol. Lett. 12, 731–738. doi: 10.1021/acs.estlett.8b00597
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Cocci, P., Mosconi, G., Bracchetti, L., Nalocca, J. M., Frapiccini, E., Marini, M., et al. (2018). Investigating the potential impact of polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs) on gene biomarker expression and global DNA methylation in loggerhead sea turtles (Caretta caretta) from the Adriatic Sea. Sci. Total Environ. 61, 49–57. doi: 10.1016/j.scitotenv.2017.11.118
Cocci, P., Mosconi, G., and Palermo, F. A. (2019). Gene expression profiles of putative biomarkers in juvenile loggerhead sea turtles (Caretta caretta) exposed to polycyclic aromatic hydrocarbons. Environ. Pollut. 246, 99–106. doi: 10.1016/j.envpol.2018.11.098
Cocci, P., Mosconi, G., and Palermo, F. A. (2020). Sunscreen active ingredients in loggerhead turtles (Caretta caretta) and their relation to molecular markers of inflammation, oxidative stress and hormonal activity in wild populations. Mar. Pollut. Bull. 153:111012. doi: 10.1016/j.marpolbul.2020.111012
Costello, E. K., Gordon, J. I., Secor, S. M., and Knight, R. (2010). Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 4, 1375–1385. doi: 10.1038/ismej.2010.71
Deng, Y., Guo, X., Wang, Y., He, M., Ma, K., Wang, H., et al. (2015). Terrisporobacter petrolearius sp. nov., isolated from an oilfield petroleum reservoir. Int. J. Syst. Evol. Microbiol. 65, 3522–3526. doi: 10.1099/ijsem.0.000450
Derraik, J. G. B. (2002). The pollution of the marine environment by plastic debris: a review. Mar. Pollut. Bull. 44, 842–852. doi: 10.1016/S0025-326X(02)00220-5
Digka, N., Bray, L., Tsangaris, C., Andreanidou, K., Kasimati, E., Kofidou, E., et al. (2020). Evidence of ingested plastics in stranded loggerhead sea turtles along the Greek coastline, East Mediterranean Sea. Environ. Pollut. 263:114596. doi: 10.1016/j.envpol.2020.114596
D’Souza, J. M., Windsor, F. M., Santillo, D., and Ormerod, S. J. (2020). Food web transfer of plastics to an apex riverine predator. Glob. Chang. Biol. 26, 3846–3857. doi: 10.1111/gcb.15139
Duan, H., Yu, L., Tian, F., Zhai, Q., Fan, L., and Chen, W. (2020). Gut microbiota: a target for heavy metal toxicity and a probiotic protective strategy. Sci. Tot. Environ. 742:140429. doi: 10.1016/j.scitotenv.2020.140429
Duangurai, T., Siengsanan-Lamont, J., Bumrungpun, C., Kaewmongkol, G., Areevijittrakul, L., Sirinarumitr, T., et al. (2019). Identification of uncultured bacteria from abscesses of exotic pet animals using broad-range nested 16S rRNA polymerase chain reaction and Sanger sequencing. Vet. World. 12, 1546–1553. doi: 10.14202/vetworld.2019.1546-1553
Duncan, E. M., Arrowsmith, J., Bain, C., Broderick, A. C., Lee, J., Metcalfe, K., et al. (2018). The true depth of the Mediterranean plastic problem: extreme microplastic pollution on marine turtle nesting beaches in Cyprus. Mar. Pollut. Bull. 136, 334–340. doi: 10.1016/j.marpolbul.2018.09.019
Duncan, E. M., Broderick, A. C., Fuller, W. J., Galloway, T. S., Godfrey, M. H., Hamann, M., et al. (2019). Microplastic ingestion ubiquitous in marine turtles. Glob. Chang. Biol. 25, 744–752. doi: 10.1111/gcb.14519
ECHA (2019). Proposals to Identify Substances of Very High Concern on the Basis of Criteria Set Out in REACH Article 57. Available online at: https://echa.europa.eu/documents/10162/1e516c08-d91e-6da3-87f7-cc0679135422 (accessed December 1, 2020).
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
European Parliament (2019). Briefing: Single-use Plastics and Fishing Gear. Available online at: https://www.europarl.europa.eu/RegData/etudes/BRIE/2018/625115/EPRS_BRI%282018%29625115_EN.pdf (accessed December 1, 2020).
Evariste, L., Barret, M., Mottier, A., Mouchet, F., Gauthier, L., and Pinelli, E. (2019). Gut microbiota of aquatic organisms: a key endpoint for ecotoxicological studies. Environ. Pollut. 248, 989–999. doi: 10.1016/j.envpol.2019.02.101
Fackelmann, G., and Sommer, S. (2019). Microplastics and the gut microbiome: how chronically exposed species may suffer from gut dysbiosis. Mar. Pollut. Bull. 143, 193–203. doi: 10.1016/j.marpolbul.2019.04.030
Farrell, P., and Nelson, K. (2013). Trophic level transfer of microplastic: Mytilus edulis (L.) to Carcinus maenas (L.). Environ. Pollut. 177, 1–3. doi: 10.1016/j.envpol.2013.01.046
Foti, M., Giacopello, C., Bottari, T., Fisichella, V., Rinaldo, D., and Mammina, C. (2009). Antibiotic Resistance of Gram Negatives isolates from loggerhead sea turtles (Caretta caretta) in the central Mediterranean Sea. Mar. Pollut. Bull. 58, 1363–1366. doi: 10.1016/j.marpolbul.2009.04.020
Franzellitti, S., Canesi, L., Auguste, M., Wathsala, R. H. G. R., and Fabbri, E. (2019). Microplastic exposure and effects in aquatic organisms: a physiological perspective. Environ. Toxicol. Pharmacol. 68, 37–51. doi: 10.1016/j.etap.2019.03.009
Franzellitti, S., Locatelli, C., Gerosa, G., Vallini, C., and Fabbri, E. (2004). Heavy metals in tissues of loggerhead turtles (Caretta caretta) from the northwestern Adriatic Sea. Comp. Biochem. Physiol. Part C T 138, 187–194. doi: 10.1016/j.cca.2004.07.008
Galgani, F., Claro, F., Depledge, M., and Fossi, C. (2014). Monitoring the impact of litter in large vertebrates in the Mediterranean Sea within the European Marine Strategy Framework Directive (MSFD): constraints, specificities and recommendations. Mar. Environ. Res. 100, 3–9. doi: 10.1016/j.marenvres.2014.02.003
Gall, S. C., and Thompson, R. C. (2015). The impact of debris on marine life. Mar. Pollut. Bull. 92, 170–179. doi: 10.1016/j.marpolbul.2014.12.041
Godon, J. J., Arulazhagan, P., Steyer, J. P., and Hamelin, J. (2016). Vertebrate bacterial gut diversity: Size also matters. BMC Ecol. 16:12. doi: 10.1186/s12898-016-0071-2
Gu, W., Liu, S., Chen, L., Liu, Y., Gu, C., Ren, H. Q., et al. (2020). Single-Cell RNA sequencing reveals size-dependent effects of polystyrene microplastics on immune and secretory cell populations from zebrafish intestines. Environ. Sci. Technol. 54, 3417–3427. doi: 10.1021/acs.est.9b06386
Hoarau, L., Ainley, L., Jean, C., and Ciccione, S. (2014). Ingestion and defecation of marine debris by loggerhead sea turtles, Caretta caretta, from by-catches in the South-West Indian Ocean. Mar. Pollut. Bull. 84, 90–96. doi: 10.1016/j.marpolbul.2014.05.031
Hooper, L. V., Littman, D. R., and Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi: 10.1126/science.1223490
Hudak, C. A., and Sette, L. (2019). Opportunistic detection of anthropogenic micro debris in harbor seal (Phoca vitulina vitulina) and gray seal (Halichoerus grypus atlantica) fecal samples from haul-outs in southeastern Massachusetts, USA. Mar. Pollut. Bull. 145, 390–395. doi: 10.1016/j.marpolbul.2019.06.020
Isangedighi, I. A., David, G. S., and Obot, O. I. (2018). Plastic waste in the aquatic environment: impacts and management. Environment 2, 1–31. doi: 10.31058/j.envi.2018.21001
Itoh, H., Tago, K., Hayatsu, M., and Kikuchi, Y. (2018). Detoxifying symbiosis: microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat. Prod. Rep. 35, 434–454. doi: 10.1039/c7np00051k
Jambeck, J. R., Geyer, R., Wilcox, C., Siegler, T. R., Perryman, M., Andrady, A., et al. (2015). Plastic waste inputs from land into the ocean. Science 347, 768–771. doi: 10.1126/science.1260352
Jin, Y., Lu, L., Tu, W., Luo, T., and Fu, Z. (2019). Impacts of polystyrene microplastic on the gut barrier, microbiota and metabolism of mice. Sci. Total Environ. 649, 308–317. doi: 10.1016/j.scitotenv.2018.08.353
Keswani, A., Oliver, D. M., Gutierrez, T., and Quilliam, R. S. (2016). Microbial hitchhikers on marine plastic debris: Human exposure risks at bathing waters and beach environments. Mar. Environ. Res. 118, 10–19. doi: 10.1016/j.marenvres.2016.04.006
Kühn, S., Bravo Rebolledo, E. L., and Van Franeker, J. A. (2015). “Deleterious effects of litter on marine life,” in Marine Anthropogenic Litter, eds M. Bergmann, L. Gutow, and M. Klages (Cham: Springer).
Kurchaba, N., Cassone, B. J., Northam, C., Ardelli, B. F., and LeMoine, C. M. R. (2020). Effects of mp polyethylene microparticles on microbiome and inflammatory response of larval zebrafish. Toxics 8:55. doi: 10.3390/toxics8030055
Le Guen, C., Suaria, G., Sherley, R. B., Ryan, P. G., Aliani, S., Boehme, L., et al. (2020). Microplastic study reveals the presence of natural and synthetic fibres in the diet of King Penguins (Aptenodytes patagonicus) foraging from South Georgia. Environ. Int. 134:105303. doi: 10.1016/j.envint.2019.105303
Li, L. L., Amara, R., Souissi, S., Dehaut, A., Duflos, G., and Monchy, S. (2020). Impacts of microplastics exposure on mussel (Mytilus edulis) gut microbiota. Sci. Total Environ. 745:141018. doi: 10.1016/j.scitotenv.2020.141018
Lin, M., Zeng, C. X., Jia, X. Q., Zhai, S. W., Li, Z. Q., and Ma, Y. (2019). The composition and structure of the intestinal microflora of Anguilla marmorata at different growth rates: a deep sequencing study. J. Appl. Microbiol. 126, 1340–1352. doi: 10.1111/jam.14174
Llorca, M., Álvarez-Muñoz, D., Ábalos, M., Rodríguez-Mozaz, S., Santos, L. H. M. L. M., León, V. M., et al. (2020). Microplastics in Mediterranean coastal area: toxicity and impact for the environment and human health. Trends Environ. Anal. Chem. 27:e00090. doi: 10.1016/j.teac.2020.e00090
Looft, T., Levine, U. Y., and Stanton, T. B. (2013). Cloacibacillus porcorum sp. nov., a mucin-degrading bacterium from the swine intestinal tract and emended description of the genus Cloacibacillus. Int. J. Syst. Evol. Microbiol. 63(Pt 6), 1960–1966. doi: 10.1099/ijs.0.044719-0
López-Martínez, S., Morales-Caselles, C., Kadar, J., and Rivas, M. L. (2020). Overview of global status of plastic presence in marine vertebrates. Glob. Change Biol. 27, 728–737. doi: 10.1111/gcb.15416
Lu, L., Wan, Z., Luo, T., Fu, Z., and Jin, Y. (2018). Polystyrene microplastics induce gut microbiota dysbiosis and hepatic lipid metabolism disorder in mice. Sci. Total Environ. 631-632, 449–458. doi: 10.1016/j.scitotenv.2018.03.051
Lucchetti, A., and Sala, A. (2010). An overview of loggerhead sea turtle (Caretta caretta) bycatch and technical mitigation measures in the Mediterranean Sea. Rev. Fish Biol. Fish. 20, 141–161. doi: 10.1007/s11160-009-9126-1
Lucchetti, A., Vasapollo, C., and Virgili, M. (2017). An interview-based approach to assess sea turtle bycatch in Italian waters. PeerJ 5:e3151. doi: 10.7717/peerj.3151
Márquez, M. R. (1990). FAO Species Catalogue. Vol.11: Sea Turtles of the World. An Annotated and Illustrated Catalogue of Sea Turtle Species Known to Date. FAO Fisheries Synopsis. No. 125. Rome: FAO.
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31
Matiddi, M., de Lucia, G. A., Silvestri, C., Darmon, G., Tomás, J., Pham, C. K., et al. (2019). Data collection on marine litter ingestion in sea turtles and thresholds for good environmental status. J. Vis. Exp. 147:59466. doi: 10.3791/59466
Matiddi, M., Hochsheid, S., Camedda, A., Baini, M., Cocumelli, C., Serena, F., et al. (2017). Loggerhead sea turtles (Caretta caretta): a target species for monitoring litter ingested by marine organisms in the Mediterranean Sea. Environ. Pollut. 230, 199–209. doi: 10.1016/j.envpol.2017.06.054
MSFD Technical Subgroup on Marine Litter Group, Hanke, F. Galgani, Werner, S., Oosterbaan, L., Nilsson, P., Fleet, D., et al. (2013). Guidance on Monitoring of Marine Litter in European Seas. Luxembourg: Publications Office of the European Union.
Nelms, S. E., Galloway, T. S., Godley, B. J., Jarvis, D. S., and Lindeque, P. K. (2018). Investigating microplastic trophic transfer in marine top predators. Environ. Pollut. 238, 999–1007. doi: 10.1016/j.envpol.2018.02.016
Nicolau, L., Marçalo, A., Ferreira, M., Sá, S., Vingada, J., and Eira, C. (2016). Ingestion of marine litter by loggerhead sea turtles, Caretta caretta, in Portuguese continental waters. Mar. Pollut. Bull. 103, 179–185. doi: 10.1016/j.marpolbul.2015.12.021
Olsen, I. (2014). “The Family Fusobacteriaceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer).
Pasquini, G., Ronchi, F., Strafella, P., Scarcella, G., and Fortibuoni, T. (2016). Seabed litter composition, distribution and sources in the Northern and Central Adriatic Sea (Mediterranean). Waste Manag. 58, 41–51. doi: 10.1016/j.wasman.2016.08.038
Peng, J., Wang, J., and Cai, L. (2017). Current understanding of microplastics in the environment: occurrence, fate, risks, and what we should do. Integr. Environ. Assess. Manag. 13, 476–482. doi: 10.1002/ieam.1912
Pham, C. K., Rodríguez, Y., Dauphin, A., Carriço, R., Frias, J. P. G. L., Vandeperre, F., et al. (2017). Plastic ingestion in oceanic-stage loggerhead sea turtles (Caretta caretta) off the North Atlantic subtropical gyre. Mar. Pollut. Bull. 121, 222–229. doi: 10.1016/j.marpolbul.2017.06.008
PlasticsEurope (2019). An Analysis of European Plastics Production, Demand and Waste Data. Available at https://www.plasticseurope.org/it/resources/publications/1804-plastics-facts-2019 (accessed December 1, 2020).
Qi, X. Z., Xue, M. Y., Yang, S. B., Zha, J. W., Wang, G. X., and Ling, F. (2017). Ammonia exposure alters the expression of immune-related and antioxidant enzymes-related genes and the gut microbial community of crucian carp (Carassius auratus). Fish Shellfish Immunol. 70, 485–492. doi: 10.1016/j.fsi.2017.09.043
Qiao, R., Sheng, C., Lu, Y., Zhang, Y., Ren, H., and Lemos, B. (2019). Microplastics induce intestinal inflammation, oxidative stress, and disorders of metabolome and microbiome in zebrafish. Sci. Total Environ. 662, 246–253. doi: 10.1016/j.scitotenv.2019.01.245
Renzi, M., Guerranti, C., and Blaškoviæ, A. (2018). Microplastic contents from maricultured and natural mussels. Mar. Pollut. Bull. 131, 248–251. doi: 10.1016/j.marpolbul.2018.04.035
Reynolds, C., and Ryan, P. G. (2018). Micro-plastic ingestion by waterbirds from contaminated wetlands in South Africa. Mar. Pollut. Bull. 126, 330–333. doi: 10.1016/j.marpolbul.2017.11.021
Rios, L. M., Moore, C., and Jones, P. R. (2007). Persistent organic pollutants carried by synthetic polymers in the ocean environment. Mar. Pollut. Bull. 54, 1230–1237. doi: 10.1016/j.marpolbul.2007.03.022
Rocha-Santos, T., and Duarte, A. C. (2015). A critical overview of the analytical approaches to the occurrence, the fate and the behavior of microplastics in the environment. TrAC Trends Anal. Chem. 65, 47–53. doi: 10.1016/j.trac.2014.10.011
Rochman, C. M., Tahir, A., Williams, S. L., Baxa, D. V., Lam, R., Miller, J. T., et al. (2015). Anthropogenic debris in seafood: plastic debris and fibers from textiles in fish and bivalves sold for human consumption. Sci. Rep. 5:14340. doi: 10.1038/srep14340
Sakamoto, M., Iino, T., and Ohkuma, M. (2017). Faecalimonas umbilicata gen. nov., sp. nov., isolated from human faeces, and reclassification of Eubacterium contortum, Eubacterium fissicatena and Clostridium oroticum as Faecalicatena contorta gen. nov., comb. nov., Faecalicatena fissicatena comb. nov. and Faecalicatena orotica comb. nov. Int. J. Syst. Evol. Microbiol. 67, 1219–1227. doi: 10.1099/ijsem.0.001790
Santos, R. G., Andrades, R., Demetrio, G. R., Kuwai, G. M., Sobral, M. F., Vieira, J. S., et al. (2020). Exploring plastic-induced satiety in foraging green turtles. Environ. Pollut. 265(Pt B):114918. doi: 10.1016/j.envpol.2020.114918
Schuyler, Q., Hardesty, B. D., Wilcox, C., and Townsend, K. (2014a). Global analysis of anthropogenic debris ingestion by sea turtles. Conserv. Biol. 28:129e139. doi: 10.1111/cobi.12126
Schuyler, Q. A., Wilcox, C., Townsend, K., Hardesty, B. D., and Marshall, N. J. (2014b). Mistaken identity? Visual similarities of marine debris to natural prey items of sea turtles. BMC Ecol. 14:14. doi: 10.1186/1472-6785-14-14
Schuyler, Q. A., Wilcox, C., Townsend, K. A., Wedemeyer-Strombel, K. R., Balazs, G., van Sebille, E., et al. (2016). Risk analysis reveals global hotspots for marine debris ingestion by sea turtles. Glob. Chang. Biol. 2, 567–576. doi: 10.1111/gcb.13078
Schwabl, P., Koppel, S., Konigshofer, P., Bucsics, T., Trauner, M., Reiberger, T., et al. (2019). Detection of various microplastics in human stool: a prospective case series. Ann. Intern. Med. 171, 453–457. doi: 10.7326/M19-0618
Semova, I., Carten, J. D., Stombaugh, J., MacKey, L. C., Knight, R., Farber, S. A., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288. doi: 10.1016/j.chom.2012.08.003
Tsuchiya, C., Sakata, T., and Sugita, H. (2008). Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 46, 43–48. doi: 10.1111/j.1472-765X.2007.02316.x
UNEP (2014). UNEP Year Book 2014 Emerging Issues Update. Nairobi: United Nations Environment Programme.
Valente, T., Sbrana, A., Scacco, U., Jacomini, C., Bianchi, J., Palazzo, L., et al. (2019). Exploring microplastic ingestion by three deep-water elasmobranch species: a case study from the Tyrrhenian Sea. Environ. Pollut. 253, 342–350. doi: 10.1016/j.envpol.2019.07.001
Wilcox, C., Puckridge, M., Schuyler, Q. A., Townsend, K., and Hardesty, B. D. (2018). A quantitative analysis linking sea turtle mortality and plastic debris ingestion. Sci. Rep. 8:12536. doi: 10.1038/s41598-018-30038-z
Keywords: loggerhead sea turtles, plastic litter, microbiome, Mediterranean Sea, plastic pollution
Citation: Biagi E, Musella M, Palladino G, Angelini V, Pari S, Roncari C, Scicchitano D, Rampelli S, Franzellitti S and Candela M (2021) Impact of Plastic Debris on the Gut Microbiota of Caretta caretta From Northwestern Adriatic Sea. Front. Mar. Sci. 8:637030. doi: 10.3389/fmars.2021.637030
Received: 02 December 2020; Accepted: 01 February 2021;
Published: 19 February 2021.
Edited by:
Monique Mancuso, National Research Council (CNR), ItalyReviewed by:
Francisco Javier Aznar, University of Valencia, SpainTeresa Bottari, Istituto per l’Ambiente Marino Costiero (IAMC), Italy
Erica Marchiori, University of Padua, Italy
Copyright © 2021 Biagi, Musella, Palladino, Angelini, Pari, Roncari, Scicchitano, Rampelli, Franzellitti and Candela. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Candela, marco.candela@unibo.it
 Elena Biagi
Elena Biagi Margherita Musella
Margherita Musella Giorgia Palladino
Giorgia Palladino Valeria Angelini
Valeria Angelini Sauro Pari3
Sauro Pari3  Simone Rampelli
Simone Rampelli Silvia Franzellitti
Silvia Franzellitti Marco Candela
Marco Candela