A de novo Transcriptome Assembly of the European Flounder (Platichthys flesus): The Preselection of Transcripts Encoding Active Forms of Enzymes
- Department of Genetics and Marine Biotechnology, Institute of Oceanology, Polish Academy of Sciences, Sopot, Poland
The RNA sequencing data sets available for different fish species show a potentially high variety of forms of enzymes just in teleosts. This is primarily considered an effect of the first round of whole-genome duplication with mutations in duplicated genes (isozymes) and alternative splicing of mRNA (isoforms). However, the abundance of the mRNA transcript variants is not necessarily reflected in the abundance of active forms of proteins. We have investigated the transcriptional profiles of two enzymes, aralkylamine N-acetyltransferase (AANAT: EC 2.3.1.87) and N-acetylserotonin O-methyltransferase (ASMT: EC 2.1.1.4), in the eyeball, brain, intestines, spleen, heart, liver, head kidney, gonads, and skin of the European flounder (Platichthys flesus). High-throughput next-generation sequencing technology NovaSeq6000 was used to generate 500M sequencing reads. These were then assembled and filtered producing 75k reliable contigs. Gene ontology (GO) terms were assigned to the majority of annotated contigs/unigenes based on the results of PFAM, PANTHER, UniProt, and InterPro protein database searches. BUSCOs statistics for metazoa, vertebrata, and actinopterygii databases showed that the reported transcriptome represents a high level of completeness. In this article, we show how to preselect transcripts encoding the active enzymes (isozymes or isoforms), using AANAT and ASMT in the European flounder as the examples. The data can be used as a tool to design the experiments as well as a basis for discussion of diversity of enzyme forms and their physiological relevance in teleosts.
The Basics
The presence of multiple forms of enzymes in teleost fish is primarily considered a result of the first round of whole-genome duplication and mutations in duplicated genes that occurred in teleostean evolutionary history (isozymes) as well as it is generated by the process of alternative splicing of mRNA (isoforms). A common occurrence of gene duplication and the scale and importance of this phenomenon are shown by Zhang (2003). Just for the record, isozymes are different forms of the same enzyme coming from different genes but catalyzing the same chemical reaction; isoforms originate from the same gene, but they can have the same or unique functions, depending on how they are spliced. Such abundance of the mRNA transcript variants as is shown in teleost fish is not necessarily reflected in the abundance of active forms of enzymes. Unfortunately, the analysis of transcriptome sequencing by de novo assembly without any mechanisms of data preselection may lead to an overestimation of the number of transcripts, and only some of them correspond to active enzymes. Although genome-based transcriptome analyses should be immune to the problem, they do require a reasonably complete genome project, which is often not available for the target species. Reasonably deep RNA-Seq is relatively easy to perform, and de novo assembly remains a method of choice for these species. Thus, a question arises as to how to effectively preselect cDNA transcripts corresponding to proteins that are active in the cells. In this article, we show it on the example of two enzymes: aralkylamine N-acetyltransferase (AANAT: EC 2.3.1.87) and N-acetylserotonin O-methyltransferase (ASMT: EC 2.1.1.4) in the European flounder (Platichthys flesus). We have investigated the transcriptional profiles of the enzymes in the eyeball, brain, intestines, spleen, heart, liver, head kidney, gonads, and skin. A link between aanat and asmt expression and the physiological role of AANAT and ASMT in various organs in fish is discussed in our previous papers (Kulczykowska et al., 2017; Pomianowski et al., 2020).
Why AANAT and ASMT?
Both AANAT and ASMT are well recognized in fish as enzymes determining the overall biosynthesis rate of melatonin (Mel; N-acetyl-5-methoxytryptamine). Mel, as a multifunctional hormone synthetized in various tissues/organs and implicated in a wide spectrum of physiological and behavioral events, has been attracting the attention of researchers for many years, the same as the enzymes controlling its synthesis in fish cells (for review, see Falcón et al., 2010, 2011, 2014; Kulczykowska et al., 2017; Pomianowski et al., 2020). For the record, AANAT converts serotonin (5-hydroxytryptamine; 5-HT) to N-acetylserotonin (NAS), and ASMT, previously named hydroxyindole-O-methyltransferase (HIOMT), methylates NAS to Mel in vertebrates, including fish. Furthermore, AANAT and ASMT are active in many other metabolic pathways: AANAT acetylates dopamine to N-acetyldopamine in fish (Zilberman-Peled et al., 2006; Paulin et al., 2015), and ASMT methylates NAS to 5-HT (and 5-HT metabolites) to 5-methoxytryptamine (5-MTAM), 5-methoxyindole acetic acid (5-MIAA), and 5-methoxytryptophol (5-MTOL) in mammals (Pévet et al., 1981; Morton, 1987).
In teleost fish, in contrast to tetrapods, there are several AANAT and ASMT isozymes encoded by distinct genes as a result of genome duplication (Falcón et al., 2009, 2011). The first report of AANAT encoding genes shows two (aanat1 and aanat2) genes expressed in pike (Esox lucius) (Coon et al., 1999). Later papers report expression of three genes: aanat1a, aanat1b (also known as snat), and aanat2 in the pufferfish (Takifugu rubripes and Tetraodon nigroviridis), medaka (Oryzias latipes) (Coon and Klein, 2006), and sea bass (Dicentrarchus labrax) (Paulin et al., 2015). Additionally, ASMT encoding genes asmt and asmt2 (also known as hiomt and hiomt2) and their transcripts or only one asmt transcript have been detected in various fish species (Velarde et al., 2010; Khan et al., 2016; Muñoz-Pérez et al., 2016; Zhang et al., 2017). Our research group has examined the expression of the aanat and asmt genes in various organs of the three-spined stickleback (Gasterosteus aculeatus) as a part of the project on the cutaneous stress response system in fish (Kulczykowska, 2019). We have found two transcripts of genes encoding AANAT and two of ASMT in the eyeball and one of each in the skin (Pomianowski et al., 2020). However, in an earlier study (Kulczykowska et al., 2017), also in the three-spined stickleback, we even found three aanats mRNAs in the brain, eye, skin, stomach, gut, heart, and kidney, but their levels differed significantly within and among organs.
A variety of AANAT and ASMT isozymes and isoforms in teleost fish detected by the analysis of transcriptome sequencing data (for example, Li et al., 2015; Zhang et al., 2017; Lv et al., 2020) together with the results of our previous studies (Kulczykowska et al., 2017; Pomianowski et al., 2020) show some discrepancies. Thus, a question arises if all transcript variants correspond to active forms of enzymes (isozymes and/or isoforms) in fish organs/tissues. The diverse properties of the encoded proteins with respect to their enzymatic activity, including substrate preferences, kinetic characteristics, and mechanism of regulation as well as their organ distribution can indicate multiple biological functions. Furthermore, different AANAT and ASMT variants (isozymes and isoforms) and their combinations, which are organ specific, can be engaged in regulation of homeostasis of the organism under different conditions and in different phases of organism development having a marked impact on fish physiology. Therefore, a preselection of transcripts corresponding to the enzymes that are active in studied organs is required. Biological importance of AANAT and ASMT resulting from their role in many metabolic pathways in the cells explains a continuous need for research on them. This paper follows this trend.
Why the European Flounder? Is It Just Another Species Examined?
The European flounder inhabits the European coastal waters from the White Sea in the north to the Mediterranean and the Black Sea in the south. The exceptional adaptability of this species to live, breed, and prosper in waters of different salinity and temperature makes it an excellent model organism in which to study various physiological processes, including osmoregulation and adaptation to variable oxygen conditions (for example, Warne and Balment, 1995, 1997; Kulczykowska et al., 2001; Lundgreen et al., 2008). Furthermore, the flounder is widely used in many studies as a bio-indicator (Hylland et al., 1996; Grinwis et al., 2000; Napierska et al., 2009; Laroche et al., 2013). This flatfish is generally readily chosen as an experimental subject as a species easily adaptable to laboratory conditions. Despite this, so far, there are no comprehensive transcriptome data from different organs of this species except for a mixed-tissue RNA-seq data set (SRX893920) released in 2015 but not described in any formal publication. The only transcriptomic and genomic data for closely related species are limited to Japanese flounder Paralichthys olivaceus (Pleuronectiformes) inhabiting the Western Pacific, which hampers any comparative molecular approaches between flatfish species so that we want to fill the gap.
It is becoming increasingly apparent that transcriptome data of the European flounder can provide new tools for study of the molecular mechanisms underlying the disruption of homeostasis, which can affect the reproductive success of fish. Therefore, such data are important not only for basic science, but they can be applied in fisheries and resource management. We found the European flounder to be a good model organism for studying the diversity of enzyme forms and potential physiological consequences of this phenomenon. We address our study to marine and freshwater biologists and ecologists, aquaculturists, toxicologists, and climatologists.
Methods
RNA Extraction and Sequencing
One European flounder (Platichthys flesus) female was collected in the Gulf of Gdańsk (Poland), transported to the Institute of Oceanology PAS, and kept in a 200-L aerated aquarium (at 7 ppt salinity, 8 ± 0.2°C water temperature and 8L:16D natural photoperiod) 2 weeks before sampling. The fish was sacrificed at 10 pm by cutting a spinal cord. Whole organs: eyeball, brain, and approximately 5 × 5 mm samples of intestine, spleen, heart, liver, head kidney, and gonad as well as skin from the upper and bottom parts of the fish were dissected immediately after sacrificing. All tissue samples were transferred to Eppendorf tubes and snap frozen in a dry ice—95% EtOH cooling bath. Frozen samples were stored at −70°C until RNA extraction. Total RNA was purified with a GenEluteTM Mammalian Total RNA Miniprep Kit (RTN70, Sigma-Aldrich, St. Louis, MO, United States) with minor modifications according to Pomianowski et al. (2020). RNA integrity number (RIN) was determined with a 2100 Bioanalyzer (Agilent), and samples with a total RNA amount ranging from 1.85 to 6.36 μg, RIN 6.5 to 8.6, and rRNA ratio 1.0 to 1.5 were used to construct sequencing libraries. Equal amounts of RNA isolates were sequenced on an Illumina NovaSeq6000 platform (TruSeq NGS library) with 150 bp paired-end run mode and 40M reads per sample throughput (Macrogen Inc., Korea). Initially, a total of more than 5.1 × 108 raw PE reads were obtained from all libraries. Then, after filtering by removal of adaptor sequences, contaminated and poor-quality reads we obtained approximately 75 Gbp of clean data (Q20 bases > 99%).
De novo Assembly of Fish Transcriptome
The Trinity version 2.9.1 (Grabherr et al., 2011) assembler with default parameters was used to obtain de novo assembly of the combined reads from all samples. There were 350,609 contigs in this initial, highly redundant assembly. The redundancies were reduced by applying CD-HIT-EST program release v4.8.1 (Li and Godzik, 2006) with parameter −c 0.98 and by filtering off very poorly represented contigs (TPM <1.0) after mapping the raw data back at the assembly as recommended by the Trinity manual (Supplementary Table 1). Additional filtering was performed after functional annotation. Contigs matching likely contaminants (similarity >85% to inconsistent taxa) were removed by our tritoconstrictor python script available on github1. Moreover, contigs without consistently annotated open reading frames and TPM <10 in at least one sample were also removed. The final filtered assembly consisted of 75,017 contigs (or Trinity isoforms) in 37,956 unigenes (defined as Trinity “groups”). The technical quality of this assembly was assessed by transrate v1.0.3 (Smith-Unna et al., 2016) (Supplementary Table 2). The completeness of the assembly was evaluated using BUSCO pipeline version 3.0.2 (Simão et al., 2015; Supplementary Figure 1). More than 93% of the 2,586 representative vertebrate BUSCOs were present in the reported transcriptome. For the metazoa and actinopterygii BUSCOs, the statistics were also very good, suggesting that the transcriptome represents a rather high level of completeness. The relatively large fraction of duplicated BUSCO for all databases suggests that the number of alternatively assembled isoforms or assembly artifacts is still high in the final assembly.
Functional Annotation of Transcriptomes
To annotate the assembled unigenes, we searched for the homologous sequences of all isoforms in three protein databases: UniRef90 (2020/02 release) (Suzek et al., 2015), PFAM release 32.0 (Finn et al., 2010), and PANTHER release 15.0 (Thomas et al., 2003). All databases were searched on a local high-performance computer cluster. The two databases containing protein profiles (PANTHER and PFAM) were searched with hmmer2 (version 3.3), UniRef90 was searched with Mmseqs2 release 11-e1a1c (Mirdita et al., 2019), and the results were integrated according to the pipeline outlined in Supplementary Figure 2. Only database hits with bitscores higher than 20 were used to produce the final annotation. Gene ontology (GO) terms were assigned to those annotated unigenes based on the current (dated 1/1/2017) official release (Ashburner et al., 2000) using mapping files provided by UniRef and PANTHER. Additionally, based on PFAM and PANTHER signatures, some unigenes were classified according to InterPro system (Mitchell et al., 2019), and GO terms for these unigenes were also integrated. The majority (20,077) of unigenes from the reference assembly were assigned some GO terms (Supplementary Figure 3). The tritoconstrictor python script performing annotation and filtering is available on github.
Analysis of aanat and asmt Transcripts
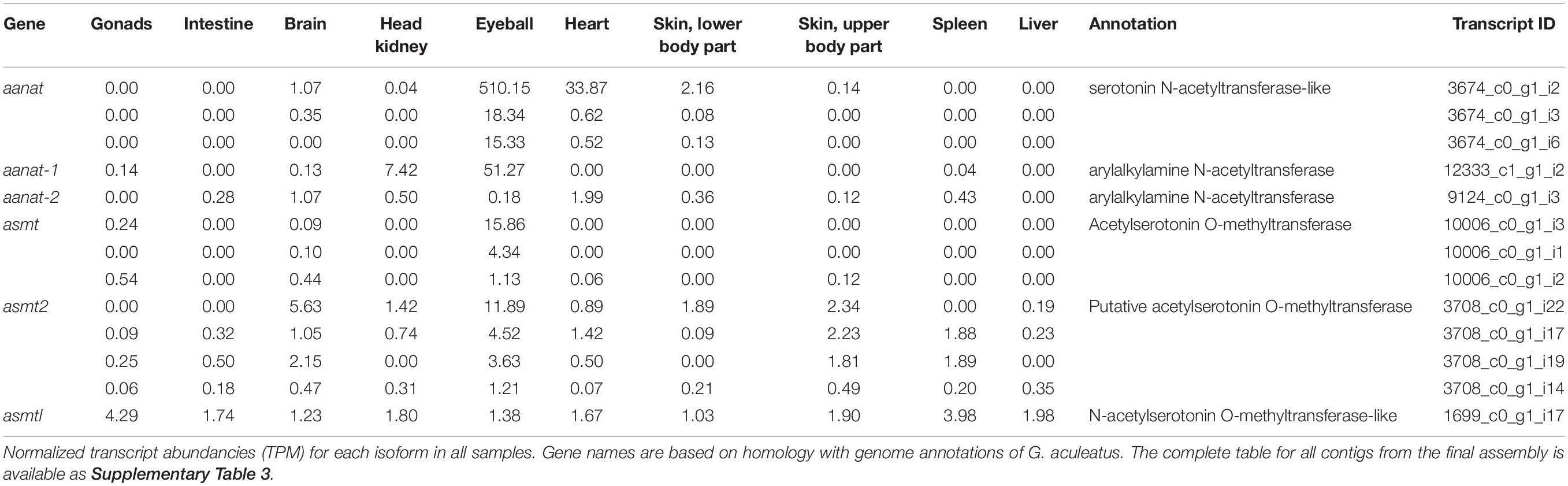
The final annotated transcriptome contained all six expected groups of transcripts of aanat and asmt, some with several potential isoforms (Table 1). The integrity of these transcripts was verified as follows. First, homologous genomic sequences of Japanese flounder (Paralichthys olivaceus) (GenBank GCA_001904815 and GCF_001970005) and stickleback (Gasterosteus aculeatus) (BROADs1, Ensemble release 97) were identified using the CLC Genomics workbench ver. 9.5.5 (Qiagen) “Large Gap Read Mapping” procedure. Then, all isoforms were aligned with Japanese flounder reference sequences. The alignments of problematic isoforms are presented in Figure 1. Based on these alignments, only a single complete transcript from each gene was identified. The remaining isoforms can be regarded as artifacts due to illegitimate assembly of disjointed sequences. The apparent presence of unspliced introns in three (out of 13) isoforms is perhaps surprising. Apparently, given the substantial sequencing depth, our RNA isolation procedure left enough genomic DNA in the sample to generate this type of assembly artifact. Despite that, the unambiguous identification of correctly spliced transcripts was possible in each case. There was no evidence for the presence of differentially expressed isoforms in any of the six genes.
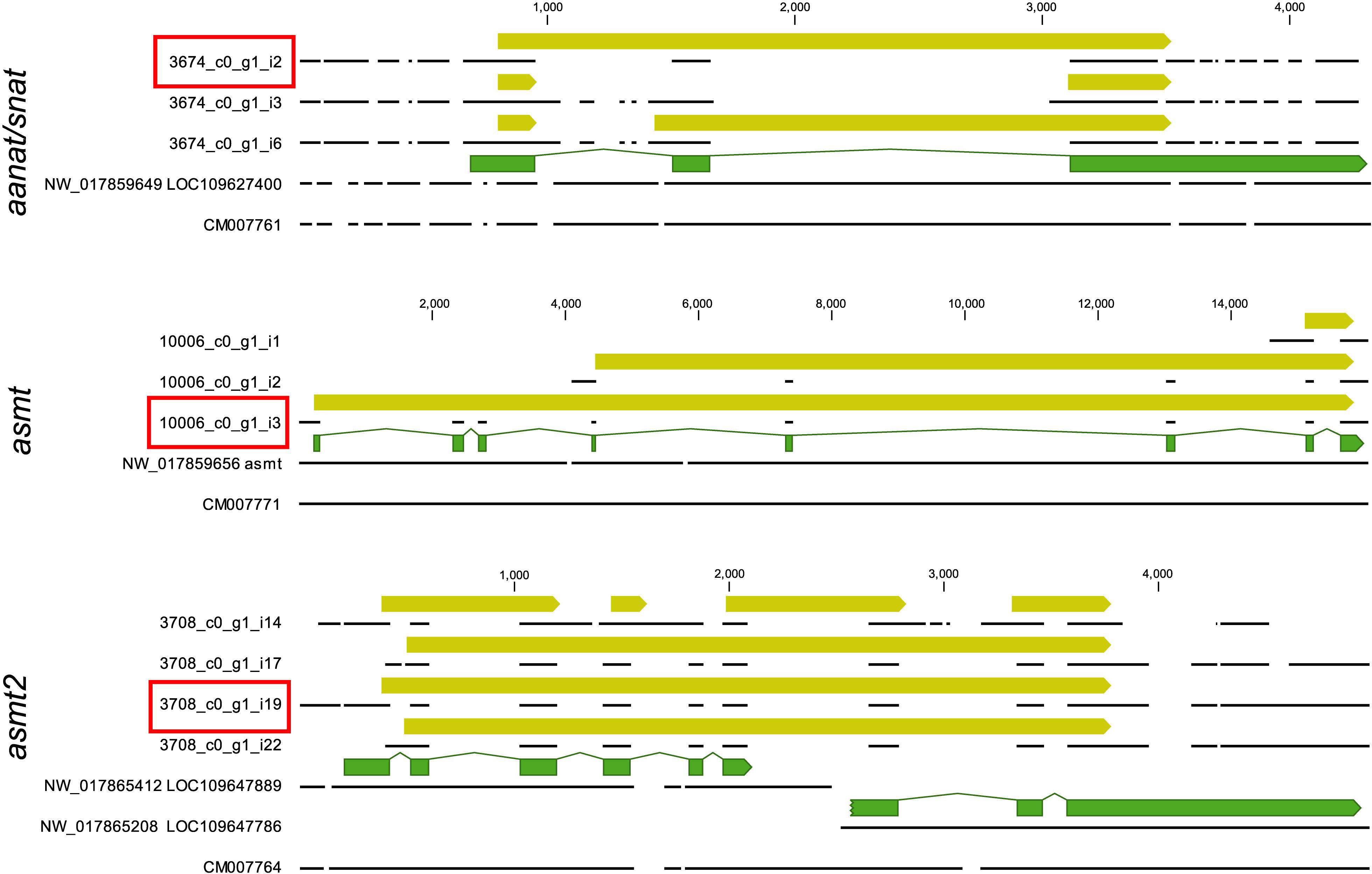
Figure 1. Alignment of aanat/snat, asmt, asmt2 transcript sequences (referenced by contig/isoform IDs used in Table 1 and Supplementary Data) to relevant genomic sequences of Japanese flounder (referenced by accession numbers). The figure was prepared in CLC Genomics Workbench and follows conventions used in this software. Annotations are presented as arrows above the sequences represented by black lines. A good match is observed only within annotated exons (green arrows), confirming the structural integrity of the annotated open reading frames (yellow arrows). Note that ORF annotations span over the alignment gaps. Only a single isoform from each locus has a complete ORF and no indication of misassembly (red boxes).
In all studied organs of the European flounder, asmtl transcripts were present at low levels (Table 1). In teleosts, asmtl gene is a product of fusion between maf and asmt genes (Zhang et al., 2017). Transcripts of the asmtl gene that were identified in Platichthys flessus had the same intron–exon structure as in the three-spined stickleback and Japanese flounder. Moreover, their sequence similarity to stickleback and Japanese flounder genomic sequences was convincing enough to conclude that these genes are truly homologous. The asmtl gene was found in the genomes of several fish species by Zhang et al. (2017). The transcriptomic survey made by the same author showed that asmtl is transcribed in the eye, skin, liver, and gonads of Sinocyclochelius fish and in the brain, gill, liver, muscle, and skin of mudskippers (Boleophthalmus pectinirosris and Periophthalmus magnuspinnatus) (Zhang et al., 2017). Taking into consideration that asmt transcripts were found mostly in the eye and brain and asmt2 and asmtl transcripts in many peripheral organs, their function seems to be different.
Data Records
The sequencing and assembly data of the transcriptome for all samples were deposited into public repositories. The raw sequencing data generated in this work were deposited in NCBI Sequence Read Archive (Leinonen et al., 2011). The assembly was deposited at DDBJ/EMBL/GenBank Transcriptome Shotgun Assembly (TSA) database. The version described in this paper is linked to NCBI BioProject number PRJNA637628.
Additional data, including expression profiling across samples, are available as a Supplementary Material (Supplementary Table 3).
Concluding Remarks
A variety of enzyme variants, isozymes and isoforms, in teleost fish, which are shown by the analysis of transcriptome sequencing data and presented in many papers, including ours, prompted us to investigate the factual diversity of the enzymes. Hence, we proposed how to effectively preselect those cDNA transcripts while analyzing transcriptome sequences to distinguish those corresponding to the active proteins. In this article, we show it on the example of two enzymes, aralkylamine N-acetyltransferase and N-acetylserotonin O-methyltransferase, in the European flounder. It is important to sequence RNA from as diverse a set of organs as possible to assure that the final transcriptome is complete enough for identification of true organs-specific alternatively spliced transcripts. Therefore, the analyses were performed in nine different organs. Discrimination of true alternatively spliced isoforms from assembly artifacts proved to be difficult based on the de novo data alone. However, raw genomic data of closely related Japanese flounder coming from two sequencing projects (GenBank GCA_001904815 and GCF_001970005) were used for final verification of identity and completeness of prefiltered isoforms. The expression of five aanat and asmt genes differed markedly between organs. Moreover, a low expression of asmtl, a gene previously described in several teleost species, was also found in nine organs of the European flounder. No compelling evidence for alternatively spliced isoforms was identified for any of the six target genes.
Data Availability Statement
This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GIPN00000000. Sample description and raw sequencing reads are deposited at NCBI databases. Accession numbers are provided in the Data column of Supplementary Table 1, and linked from the relevant BioProject site (accession number PRJNA637628).
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee for Animal Experimentation (University of Science and Technology, Bydgoszcz) Mazowiecka 28 Str. 85-084 Bydgoszcz Poland.
Author Contributions
KP and AB were the authors of the research idea. KP assisted in sample collection, extracted the RNA and estimated its quality and integrity. AB assembled and annotated the transcriptome. EK, KP, and AB contributed to writing the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Science Centre (Poland) grant UMO-2017/27/B/NZ4/01259.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Mr. Ireneusz Stepczyński for catching fish investigated in the presented research and Mrs. Hanna Kalamarz-Kubiak, Ph.D., DSc., for tissue sample collection from the investigated animal. Computer analyses in this study were run on supercomputers of the Academic Computer Centre (TASK) in Gdańsk, as well as using the PLGRID (www.plgrid.pl) infrastructure.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.618779/full#supplementary-material
Supplementary Figure 1 | BUSCO assessment of assembly completeness. The final filtered assembly (75k contigs) was compared with the three reference sets of BUSCOs as defined in www.orthodb.org database version 9. For the smallest metazoan data set, only 5 genes were not found in the assembly out of the 978 searched for, indicating good completeness of the assembly.
Supplementary Figure 2 | Simplified overview of assembly annotation and filtering workflow.
Supplementary Figure 3 | Functional annotation of the assembly. There were 37,956 unigenes in the final assembly consisting of 75,017 contigs. Out of these, 20,090 unigenes were assigned at least one Gene Ontology (GO) term. The Venn diagram shows the distribution of unigenes among the three GO categories: cellular component, biological process, and molecular function. The colored areas are proportional to the number of unigenes with respective assignments.
Supplementary Table 1 | List of raw reads and per sample assembly statistics.
Supplementary Table 2 | Transrate assembly assessment of the final assembly.
Supplementary Table 3 | The transcript expression values across selected European flounder organs. Values are presented in normalized transcripts per million (TPM) units.
Footnotes
References
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/7555
Coon, S. L., Bégay, V., Deurloo, D., Falcón, J., and Klein, D. C. (1999). Two arylalkylamine N-acetyltransferase genes mediate melatonin synthesis in fish. J. Biol. Chem. 274, 9076–9082. doi: 10.1074/jbc.274.13.9076
Coon, S. L., and Klein, D. C. (2006). Evolution of arylalkylamine N-acetyltransferase: emergence and divergence. Mol. Cell. Endocrinol. 252, 2–10. doi: 10.1016/j.mce.2006.03.039
Falcón, J., Besseau, L., Fuentès, M., Sauzet, S., Magnanou, E., and Boeuf, G. (2009). Structural and functional evolution of the pineal melatonin system in vertebrates. Ann. N.Y. Acad. Sci. 1163, 101–111. doi: 10.1111/j.1749-6632.2009.04435.x
Falcón, J., Besseau, L., Magnanou, E., Herrero, M. J., Nagai, M., and Boeuf, G. (2011). Melatonin, the timekeeper: biosynthesis and effects in fish. Cybium 35, 3–18. doi: 10.26028/cybium/2011-351-001
Falcón, J., Coon, S. L., Besseau, L., Cazaméa-Catalan, D., Fuentès, M., Magnanou, E., et al. (2014). Drastic neofunctionalization associated with evolution of the timezyme AANAT 500 Mya. Proc. Natl. Acad. Sci. U.S.A. 111, 314–319. doi: 10.1073/pnas.1312634110
Falcón, J., Migaud, H., Munoz-Cueto, J. A., and Carrillo, M. (2010). Current knowledge on the melatonin system in teleost fish. Gen. Comp. Endocrinol. 165, 469–482. doi: 10.1016/j.ygcen.2009.04.026
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., and Pollington, J. E. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Grinwis, G. C., Vethaak, A. D., Wester, P. W., and Vos, J. G. (2000). Toxicology of environmental chemicals in the flounder (Platichthys flesus) with emphasis on the immune system: field, semi-field (mesocosm) and laboratory studies. Toxicol. Lett. 112-113, 289–301. doi: 10.1016/s0378-4274(99)00239-8
Hylland, K., Sandvik, M., Utne Skare, J., Beyer, J., Egaas, E., and Goksoyr, A. (1996). Biomarkers in flounder (Platichthys flesus): an evaluation of their use in pollution monitoring. Mar. Environ. Res. 42, 223–227. doi: 10.1016/0141-1136(95)00034-8
Khan, Z. A., Yumnamcha, T., and Rajiv, C. (2016). Melatonin biosynthesizing enzyme genes and clock genes in ovary and whole brain of zebrafish (Danio rerio): differential expression and a possible interplay. Gen. Comp. Endocrinol. 233, 16–31.
Kulczykowska, E. (2019). Stress response system in the fish skin-welfare measures revisited. Front. Physiol. 10:72. doi: 10.3389/fphys.2019.00072
Kulczykowska, E., Kleszczyńska, A., Gozdowska, M., and Sokołowska, E. (2017). The time enzyme in melatonin biosynthesis in fish: day/night expressions of three aralkylamine N-acetyltransferase genes in three-spined stickleback. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 208, 46–53. doi: 10.1016/j.cbpa.2017.03.005
Kulczykowska, E., Warne, J. M., and Balment, R. J. (2001). Day-night variations in plasma melatonin and arginine vasotocin concentrations in chronically cannulated flounder (Platichthys flesus). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130, 827–834. doi: 10.1016/s1095-6433(01)00444-5
Laroche, J., Gauthier, O., Quiniou, L., Devaux, A., Bony, S., Evrard, E., et al. (2013). Variation patterns in individual fish responses to chemical stress among estuaries, seasons and genders: the case of the European flounder (Platichthys flesus) in the Bay of Biscay. Environ. Sci. Pollut. Res. 20, 738–748. doi: 10.1007/s11356-012-1276-3
Leinonen, R., Sugawara, H., Shumway, M., and International Nucleotide Sequence Database Collaboration (2011). The sequence read archive. Nucleic Acids Res. 39, D19–D21. doi: 10.1093/nar/gkq1019
Li, J., You, X., Bian, C., Yu, H., Coon, S. L., and Shi, Q. (2015). Molecular evolution of aralkylamine N-acetyltransferase in fish: a genomic survey. Int. J. Mol. Sci. 17:51. doi: 10.3390/ijms17010051
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics (Oxford, Engl.) 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Lundgreen, K., Kiilerich, P., Tipsmark, C. K., Madsen, S. S., and Jensen, F. B. (2008). Physiological response in the European flounder (Platichthys flesus) to variable salinity and oxygen conditions. J. Comp. Physiol. B 178, 909–915. doi: 10.1007/s00360-008-0281-9
Lv, Y., Li, Y., Li, J., Bian, C., Qin, C., and Shi, Q. (2020). A Comparative genomics study on the molecular evolution of serotonin/melatonin biosynthesizing enzymes in vertebrates. Front. Mol. Biosci. 7:11. doi: 10.3389/fmolb.2020.00011
Mirdita, M., Steinegger, M., and Söding, J. (2019). MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics (Oxford, Engl.) 35, 2856–2858. doi: 10.1093/bioinformatics/bty1057
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, 351–360. doi: 10.1093/nar/gky1100
Morton, D. J. (1987). Hydroxyindole-O-methyltransferase catalyses production of methoxyindoles in rat pineal gland dependent on the concentration of hydroxy precursors and their affinity for the enzyme. J. Endocrinol. 115, 455–458. doi: 10.1677/joe.0.1150455
Muñoz-Pérez, J. L., López-Patiño, M. A., Álvarez-Otero, R., Gesto, M., Soengas, J. L., and Míguez, J. M. (2016). Characterization of melatonin synthesis in the gastrointestinal tract of rainbow trout (Oncorhynchus mykiss): distribution, relation with serotonin, daily rhythms and photoperiod regulation. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 186, 471–484. doi: 10.1007/s00360-016-0966-4
Napierska, D., Barsiene, J., Mulkiewicz, E., Podolska, M., and Rybakovas, A. (2009). Biomarker responses in flounder Platichthys flesus from the Polish coastal area of the Baltic Sea and applications in biomonitoring. Ecotoxicology (London, Engl.) 18, 846–859. doi: 10.1007/s10646-009-0328-z
Paulin, C. H., Cazaméa-Catalan, D., Zilberman-Peled, B., Herrera-Perez, P., Sauzet, S., Magnanou, E., et al. (2015). Subfunctionalization of arylalkylamine N-acetyltransferases in the sea bass Dicentrarchus labrax: two-ones for one two. J. Pineal. Res. 59, 354–364. doi: 10.1111/jpi.12266
Pévet, P., Balemans, M. G., and de Reuver, G. F. (1981). The pineal gland of the mole (Talpa europaea L.). VII. Activity of hydroxyindole-O-methyltransferase (HIOMT) in the formation of 5-methoxytryptophan, 5-methoxytryptamine, 5-methoxyindole-3-acetic acid, 5-methoxytryptophol and melantonin in the eyes and the pineal gland. J. Neural Transm. 51, 271–282. doi: 10.1007/BF01248958
Pomianowski, K., Gozdowska, M., Burzyński, A., Kalamarz-Kubiak, H., Sokołowska, E., Kijewska, A., et al. (2020). A study of aanat and asmt expression in the three-spined stickleback eye and skin: not only “on the way to melatonin”. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 241:110635. doi: 10.1016/j.cbpa.2019.110635
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics (Oxford, Engl.) 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M., and Kelly, S. (2016). TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26, 1134–1144. doi: 10.1101/gr.196469.115
Suzek, B. E., Wang, Y., Huang, H., McGarvey, P. B., and Wu, C. H. (2015). UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932. doi: 10.1093/bioinformatics/btu739
Thomas, P. D., Campbell, M. J., Kejariwal, A., Mi, H., Karlak, B., Daverman, R., et al. (2003). PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141. doi: 10.1101/gr.772403
Velarde, E., Cerdá-Reverter, J. M., Alonso-Gómez, A. L., Sánchez, E., Isorna, E., and Delgado, M. J. (2010). Melatonin-synthesizing enzymes in pineal, retina, liver, and gut of the goldfish (Carassius): mRNA expression pattern and regulation of daily rhythms by lighting conditions. Chronobiol. Int. 27, 1178–1201. doi: 10.3109/07420528.2010.496911
Warne, J. M., and Balment, R. J. (1995). Effect of acute manipulation of blood volume and osmolality on plasma [AVT] in seawater flounder. Am. J. Physiol. 269(5 Pt 2), R1107–R1112. doi: 10.1152/ajpregu.1995.269.5.R1107
Warne, J. M., and Balment, R. J. (1997). Changes in plasma arginine vasotocin (AVT) concentration and dorsal aortic blood pressure following AVT injection in the teleost Platichthys flesus. Gen. Comp. Endocrinol. 105, 358–364. doi: 10.1006/gcen.1996.6837
Zhang, J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/S0169-5347(03)00033-8
Zhang, K., Ruan, Z., Li, J., Bian, C., You, X., Coon, S. L., et al. (2017). A comparative genomic and transcriptomic survey provides novel insights into N-acetylserotonin methyltransferase (ASMT) in fish. Molecules (Basel, Switzerland) 22:1653. doi: 10.3390/molecules22101653
Keywords: Aralkylamine N-acetyltransferase, N-acetylserotonin O-methyltransferase, aanat expression, asmt expression, asmtl expression, fish, isoform, isozyme
Citation: Pomianowski K, Burzyński A and Kulczykowska E (2021) A de novo Transcriptome Assembly of the European Flounder (Platichthys flesus): The Preselection of Transcripts Encoding Active Forms of Enzymes. Front. Mar. Sci. 8:618779. doi: 10.3389/fmars.2021.618779
Received: 18 October 2020; Accepted: 26 January 2021;
Published: 18 February 2021.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Vittoria Roncalli, University of Naples Federico II, ItalyChristophe Klopp, Institut National de la Recherche Agronomique de Toulouse, France
Copyright © 2021 Pomianowski, Burzyński and Kulczykowska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Konrad Pomianowski, k.pomianowski@iopan.pl
†ORCID: Konrad Pomianowski, orcid.org/0000-0002-5256-3847; Artur Burzyński, orcid.org/0000-0002-0185-197X; Ewa Kulczykowska, orcid.org/0000-0002-3979-9310
 Konrad Pomianowski
Konrad Pomianowski Artur Burzyński
Artur Burzyński Ewa Kulczykowska
Ewa Kulczykowska