Abstract
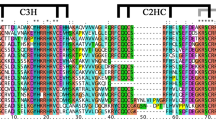
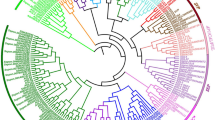
The MADS-box genes are an important class of transcription factors and play critical roles in flower development. However, the functions of these genes in the economically important drinking plant, Camellia sinensis, are still not reported. Here, an evolutionary analysis of tea MADS-box genes was performed at whole genome level. A total of 83 MADS-box genes were identified in tea, and their gene structures and expression patterns were further analyzed. The tea MADS-box genes were classified into Mα (26), Mβ (12), Mγ (9), MIKC* (7), and MIKCC (29) clade according to their phylogenetic relationship with Arabidopsis thaliana. Several cis-elements were identified in the promoter regions of the CsMADS genes that are important in regulating growth, development, light responses, and the response to several stresses. Most CsMADS genes display clear different expression patterns in different organs and different species of tea plant. The expression of CsMADS genes can be regulated by abiotic stresses and phytohormone treatment. Our results lay the foundation for future research on the function of CsMADS genes and beneficial for improving tea agricultural traits in the future.








Similar content being viewed by others
References
Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martínez-Castilla L, Yanofsky MF (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci U S A 97(10):5328–5333
Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242
Bai G, Yang DH, Cao P, Yao H, Zhang Y, Chen X, Xiao B, Li F, Wang ZY, Yang J, Xie H (2019) Genome-wide identification, gene structure and expression analysis of the MADS-box gene family indicate their function in the development of tobacco (Nicotiana tabacum L.). Int J Mol Sci 20(20)
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37(Web Server issue):W202–W208
Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112(1):1–20
Chen R, Ma J, Luo D, Hou X, Ma F, Zhang Y, Meng Y, Zhang H, Guo W (2019) CaMADS, a MADS-box transcription factor from pepper, plays an important role in the response to cold, salt, and osmotic stress. Plant Sci 280:164–174
Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14(21):1935–1940
Dreni L, Zhang D (2016) Flower development: the evolutionary history and functions of the AGL6 subfamily MADS-box genes. J Exp Bot 67(6):1625–1638
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44(D1):D279–D285
Gregis V, Sessa A, Dorca-Fornell C, Kater MM (2009) The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J 60(4):626–637
Grimplet J, Martínez-Zapater JM, Carmona MJ (2016) Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genomics 17:80
Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46(2):183–192
Khong GN, Pati PK, Richaud F, Parizot B, Bidzinski P, Mai CD, Bès M, Bourrié I, Meynard D, Beeckman T, Selvaraj MG, Manabu I, Genga AM, Brugidou C, Nang Do V, Guiderdoni E, Morel JB, Gantet P (2015) OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol 169(4):2935–2949
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33(7):1870–1874
Lai X, Daher H, Galien A, Hugouvieux V, Zubieta C (2019) Structural basis for plant MADS transcription factor oligomerization. Comput Struct Biotechnol J 17:946–953
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43(Database issue):D257–D260
Ma J, Yang Y, Luo W, Yang C, Ding P, Liu Y, Qiao L, Chang Z, Geng H, Wang P, Jiang Q, Wang J, Chen G, Wei Y, Zheng Y, Lan X (2017) Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.). PLoS One 12(7):e0181443
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res 43(Database issue):D222–D226
Mondal TK (2003) Micropropagation of tea (Camellia sinensis L.). Kluwer Publication, The Netherlands
Ó'Maoiléidigh DS, Graciet E, Wellmer F (2014) Gene networks controlling Arabidopsis thaliana flower development. New Phytol 201(1):16–30
Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15(7):1538–1551
Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424(6944):85–88
Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15(5):1159–1169
Ruelens P, Zhang Z, van Mourik H, Maere S, Kaufmann K, Geuten K (2017) The Origin of Floral Organ Identity Quartets. Plant Cell 29(2):229–242
Schmitz RJ, Sung S, Amasino RM (2008) Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci U S A 105(2):411–416
Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci U S A 97(7):3753–3758
Sheng XG, Zhao ZQ, Wang JS, Yu HF, Shen YS, Zeng XY, Gu HH (2019) Genome wide analysis of MADS-box gene family in Brassica oleracea reveals conservation and variation in flower development. BMC Plant Biol 19(1):106
Shu Y, Yu D, Wang D, Guo D, Guo C (2013) Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol Biol Rep 40(6):3901–3911
Wang P, Wang S, Chen Y, Xu X, Guang X, Zhang Y (2019) Genome-wide analysis of the MADS-box gene family in watermelon. Comput Biol Chem 80:341–350
Wei C, Yang H, Wang S, Zhao J, Liu C, Gao L, Xia E, Lu Y, Tai Y, She G, Sun J, Cao H, Tong W, Gao Q, Li Y, Deng W, Jiang X, Wang W, Chen Q, Zhang S, Li H, Wu J, Wang P, Li P, Shi C, Zheng F, Jian J, Huang B, Shan D, Shi M, Fang C, Yue Y, Li F, Li D, Wei S, Han B, Jiang C, Yin Y, Xia T, Zhang Z, Bennetzen JL, Zhao S, Wan X (2018) Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci U S A 115(18):E4151–e4158
Wellmer F, Graciet E, Riechmann JL (2014) Specification of floral organs in Arabidopsis. J Exp Bot 65(1):1–9
Wellmer F, Riechmann JL (2010) Gene networks controlling the initiation of flower development. Trends Genet 26(12):519–527
Wilkins MR, Gasteiger E, Bairoch A, Sanchez J-C, Williams KL, Appel RD, Hochstrasser DF (1999) Protein identification and analysis tools in the ExPASy server. In: Link AJ (ed) 2-D proteome analysis protocols. Humana Press, Totowa, pp 531–552
Wu Y, Ke Y, Wen J, Guo P, Ran F, Wang M, Liu M, Li P, Li J, Du H (2018) Evolution and expression analyses of the MADS-box gene family in Brassica napus. PLoS One 13(7):e0200762
Xia EH, Zhang HB, Sheng J, Li K, Zhang QJ, Kim C, Zhang Y, Liu Y, Zhu T, Li W, Huang H, Tong Y, Nan H, Shi C, Shi C, Jiang JJ, Mao SY, Jiao JY, Zhang D, Zhao Y, Zhao YJ, Zhang LP, Liu YL, Liu BY, Yu Y, Shao SF, Ni DJ, Eichler EE, Gao LZ (2017) The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant 10(6):866–877
Zhang X, Fatima M, Zhou P, Ma Q, Ming R (2020) Analysis of MADS-box genes revealed modified flowering gene network and diurnal expression in pineapple. BMC Genomics 21(1):8
Zhang X, Li L, Yang C, Cheng Y, Han Z, Cai Z, Nian H, Ma Q (2020) GsMAS1 encoding a MADS-box transcription factor enhances the tolerance to aluminum stress in Arabidopsis thaliana. Int J Mol Sci 21(6)
Funding
The authors are supported by Nanhu Scholars Program for Young Scholars of Xinyang Normal University.
Author information
Authors and Affiliations
Contributions
Z.-B.Z. designed the research. Z.-B.Z. and Y.-J.J. wrote the manuscript. Y.-J.J., H.-H.W., and Z.-G.F. performed the identification of MADS genes, protein structure, evolution analysis, and expression analysis. L.C. participated in manuscript preparation and revision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by: Izabela Pawłowicz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, ZB., Jin, YJ., Wan, HH. et al. Genome-wide identification and expression analysis of the MADS-box transcription factor family in Camellia sinensis. J Appl Genetics 62, 249–264 (2021). https://doi.org/10.1007/s13353-021-00621-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-021-00621-8