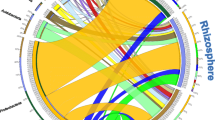
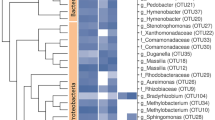
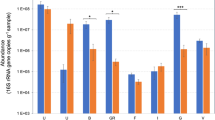
Abstract
Aims
The pea seeds (Pisum sativum) hosts a complex microbial community that is exposed to environmental constraints modulated by biotic, abiotic and anthropogenic factors. The aim of this study was to explore the effect of geographical origins on the composition of the microbial communities associated with pea seeds from storage locations in different countries. These seeds are used by Roquette Frères® as source proteins for human food after industrial extraction process of their proteins.
Methods
The high-throughput sequencing of two phylogenetic markers (V3-V4 regions of the 16S rRNA gene and ITS2 region) was used to evaluate the structure of the microbial community associated with pea seeds. The pea seeds were harvested by different suppliers from three countries (France, Sweden and Canada) with different climates and stored in six cooperatives. Representative microbial strains of dominant species were isolated and characterized using phenotypic methods.
Results
Although the pea seed microbiota shared a dominant core microbiota at the genus level, seeds showed a species-specific, highly unique microbial signature with variable abundances according to origin. Origin was identified as the main driver of the composition of the pea seed microbiota.
Conclusions
These results provide an initial insight into the broad factors such as country and cooperative that may determine the assembly of the pea microbiota before processing.







Similar content being viewed by others
Abbreviations
- BHI:
-
Brain Heart Infusion Broth
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylenediaminetetraacetic acid
- ITS:
-
Internal Transcribed Spacer
- LAB:
-
Lactic Acid Bacteria
- MRS:
-
De Man, Rogosa and Sharpe
- OTUs:
-
Operational Taxonomic Units
- RNA:
-
Ribonucleic Acid
- TTC:
-
Tetrazolium chloride
- YEGC:
-
Yeast Extract Glucose Chloramphenicol
References
Abarenkov K, Nilsson RH, Larsson KH et al (2010) The UNITE database for molecular identification of fungi - recent updates and future perspectives. New Phytol 186:281–285. https://doi.org/10.1111/j.1469-8137.2009.03160.x
Adam E, Bernhart M, Müller H, Winkler J, Berg G (2018) The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil 422:35–49. https://doi.org/10.1007/s11104-016-3113-9
Al-Awadhi H, El-Nemr I, Mahmoud H et al (2009) Plant-associated bacteria as tools for the phytoremediation of oily nitrogen-poor soils. Int J Phytoremediation 11:11–27. https://doi.org/10.1080/15226510802363261
Alexandratos N, Bruinsma J (2012) World agriculture towards 2030/2050: the 2012 revision. ESA Working paper Rome, FAO
Avis TJ, Gravel V, Antoun H, Tweddell RJ (2008) Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol Biochem 40:1733–1740. https://doi.org/10.1016/j.soilbio.2008.02.013
Bahram M, Peay KG, Tedersoo L (2015) Local-scale biogeography and spatiotemporal variability in communities of mycorrhizal fungi. New Phytol 205:1454–1463. https://doi.org/10.1111/nph.13206
Bakker MG, McCormick SP (2019) Microbial correlates of Fusarium load and Deoxynivalenol content in individual wheat kernels. Phytopathology® 109:993–1002. https://doi.org/10.1094/PHYTO-08-18-0310-R
Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, Hunault G, Simoneau P, Jacques MA (2015) Emergence shapes the structure of the seed microbiota. Appl Environ Microbiol 81:1257–1266. https://doi.org/10.1128/AEM.03722-14
Barret M, Guimbaud JF, Darrasse A, Jacques MA (2016) Plant microbiota affects seed transmission of phytopathogenic microorganisms. Mol Plant Pathol 17:791–795. https://doi.org/10.1111/mpp.12382
Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, de Wit P, Sánchez-García M, Ebersberger I, de Sousa F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. https://doi.org/10.1111/2041-210X.12073
Bhat R, Karim AA (2009) Exploring the nutritional potential of wild and underutilized legumes. Compr Rev Food Sci Food Saf 8:305–331. https://doi.org/10.1111/j.1541-4337.2009.00084.x
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. https://doi.org/10.1038/nmeth.2276
Bokulich NA, Thorngate JH, Richardson PM, Mills DA (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci India Sect B Biol Sci 111:E139–E148. https://doi.org/10.1073/pnas.1317377110
Bouchenak M, Lamri-Senhadji M (2013) Nutritional quality of legumes, and their role in cardiometabolic risk prevention: a review. J Med Food 16:185–198. https://doi.org/10.1089/jmf.2011.0238
Boye J, Zare F, Pletch A (2010) Pulse proteins: processing, characterization, functional properties and applications in food and feed. Food Res Int 43:414–431. https://doi.org/10.1016/j.foodres.2009.09.003
Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. https://doi.org/10.1146/annurev-arplant-050312-120106
Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Morsy M, Eisen JA, Leach JE, Dangl JL (2017) Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15:e2001793. https://doi.org/10.1371/journal.pbio.2001793
Chatterjee S, Sau GB, Mukherjee SK (2009) Plant growth promotion by a hexavalent chromium reducing bacterial strain, Cellulosimicrobium cellulans KUCr3. World J Microbiol Biotechnol 25:1829–1836. https://doi.org/10.1007/s11274-009-0084-5
Chesneau G, Torres-Cortes G, Briand M, Darrasse A, Preveaux A, Marais C, Jacques MA, Shade A, Barret M (2020) Temporal dynamics of bacterial communities during seed development and maturation. FEMS Microbiol Ecol:96. https://doi.org/10.1093/femsec/fiaa190
Colwell RR (2000) Viable but nonculturable bacteria: a survival strategy. J Infect Chemother 6:121–125. https://doi.org/10.1007/PL00012151
Cottyn B, Debode J, Regalado E, Mew TW, Swings J (2009) Phenotypic and genetic diversity of rice seed-associated bacteria and their role in pathogenicity and biological control. J Appl Microbiol 107:885–897. https://doi.org/10.1111/j.1365-2672.2009.04268.x
Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci 106:16428–16433. https://doi.org/10.1073/pnas.0905240106
Dietrich M, Kearney T, Seamark EC, Markotter W (2017) The excreted microbiota of bats: evidence of niche specialisation based on multiple body habitats. FEMS Microbiol Lett:364. https://doi.org/10.1093/femsle/fnw284
Duc G, Jeuffroy MH, Tivoli B (2011) Les légumineuses protéagineuses pour améliorer les bilans environnementaux en grandes cultures: principaux travaux de l’INRA qui ont accompagné la naissance de la filière et les perspectives. Innov Agronomiques 12:157–180
Dugat-Bony E, Garnier L, Denonfoux J, Ferreira S, Sarthou AS, Bonnarme P, Irlinger F (2016) Highlighting the microbial diversity of 12 French cheese varieties. Int J Food Microbiol 238:265–273. https://doi.org/10.1016/j.ijfoodmicro.2016.09.026
Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS (2019) Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol 27:105–117. https://doi.org/10.1016/j.tim.2018.11.003
Escudié F, Auer L, Bernard M, Mariadassou M, Cauquil L, Vidal K, Maman S, Hernandez-Raquet G, Combes S, Pascal G (2018) FROGS: find, rapidly, OTUs with galaxy solution. Bioinformatics 34:1287–1294. https://doi.org/10.1093/bioinformatics/btx791
Eyre AW, Wang M, Oh Y, Dean RA (2019) Identification and characterization of the Core Rice seed microbiome. Phytobiomes J. 3:148–157. https://doi.org/10.1094/PBIOMES-01-19-0009-R
Food and Agriculture Organization of the United Nations (FAO) (2020) FAOSTAT statistical database. FAO, Rome. http://www.fao.org/faostat. Accessed 12 Oct 2020
Fürnkranz M, Lukesch B, Müller H, Huss H, Grube M, Berg G (2012) Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microb Ecol 63:418–428. https://doi.org/10.1007/s00248-011-9942-4
Gangwar M, Dogra S, Sharma N (2011) Antagonistic bioactivity of endophytic actinomycetes isolated from medicinal plants. J Adv Lab Res Biol 2:154–157
Gholizadeh A, Kohnehrouz BB (2010) A molecular evidence for the presence of methylobacterial-type Fe siderophore receptor in Celosia cristata. Plant Soil Environ 56:133–138. https://doi.org/10.17221/194/2009-PSE
Glassner H, Zchori-Fein E, Yaron S, Sessitsch A, Sauer U, Compant S (2018) Bacterial niches inside seeds of Cucumis melo L. Plant Soil 422:101–113. https://doi.org/10.1007/s11104-017-3175-3
Glick BR (2012) Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012:1–15. https://doi.org/10.6064/2012/963401
Global Market Insights (2020) Pea protein market size by product (isolate, concentrate, textured), by application (meat substitutes, nutraceuticals, sports supplements), industry analysis report, regional outlook, application potential, price trends, competitive market share & forecast, 2020–2026. https://www.gminsights.com/industry-analysis/pea-protein-market-report. Accessed 12 Oct 2020
Godebo AT, Germida JJ, Walley F (2020) Isolation, identification and assessment of soil bacteria as biocontrol agents of pea root rot caused by Aphanomyces euteiches. Can J soil Sci CJSS-2019-0133. https://doi.org/10.1139/CJSS-2019-0133
Goggin DE, Emery RJN, Kurepin LV, Powles SB (2015) A potential role for endogenous microflora in dormancy release, cytokinin metabolism and the response to fluridone in Lolium rigidum seeds. Ann Bot 115:293–301. https://doi.org/10.1093/aob/mcu231
Granér G, Persson P, Meijer J, AlstrÃm S (2003) A study on microbial diversity in different cultivars of Brassica napus in relation to its wilt pathogen. Verticillium longisporum FEMS Microbiol Lett 224:269–276. https://doi.org/10.1016/S0378-1097(03)00449-X
Gundel PE, Rudgers JA, Ghersa CM (2011) Incorporating the process of vertical transmission into understanding of host-symbiont dynamics. Oikos 120:1121–1128. https://doi.org/10.1111/j.1600-0706.2011.19299.x
Gupta R, Bisaria VS, Sharma S (2015) Effect of agricultural amendments on Cajanus cajan (Pigeon pea) and its rhizospheric microbial communities–a comparison between chemical fertilizers and bioinoculants. PLoS One 10:e0132770. https://doi.org/10.1371/journal.pone.0132770
Hashidoko Y (2005) Ecochemical studies of interrelationships between epiphytic bacteria and host plants via secondary metabolites. Biosci Biotechnol Biochem 69:1427–1441. https://doi.org/10.1271/bbb.69.1427
Hu L, Robert CAM, Cadot S, Zhang X, Ye M, Li B, Manzo D, Chervet N, Steinger T, van der Heijden MGA, Schlaeppi K, Erb M (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9:2738. https://doi.org/10.1038/s41467-018-05122-7
Innerebner G, Knief C, Vorholt JA (2011) Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol 77:3202–3210. https://doi.org/10.1128/AEM.00133-11
Jain A, Singh A, Singh S, Singh HB (2015) Biological management of Sclerotinia sclerotiorum in pea using plant growth promoting microbial consortium: growth promotion effect of microbial consortium. J Basic Microbiol 55:961–972. https://doi.org/10.1002/jobm.201400628
Javaid A (2006) Foliar application of effective microorganisms on pea as an alternative fertilizer. Agron Sustain Dev 26:257–262. https://doi.org/10.1051/agro:2006024
Johnston-Monje D, Raizada MN (2011) Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One 6:e20396. https://doi.org/10.1371/journal.pone.0020396
Jumpponen A, Jones KL (2009) Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytol 184:438–448. https://doi.org/10.1111/j.1469-8137.2009.02990.x
Karthik C, Oves M, Thangabalu R, Sharma R, Santhosh SB, Indra Arulselvi P (2016) Cellulosimicrobium funkei-like enhances the growth of Phaseolus vulgaris by modulating oxidative damage under chromium(VI) toxicity. J Adv Res 7:839–850. https://doi.org/10.1016/j.jare.2016.08.007
Kecskeméti E, Berkelmann-Löhnertz B, Reineke A (2016) Are epiphytic microbial communities in the carposphere of ripening grape clusters (Vitis vinifera L.) different between conventional, organic, and biodynamic grapes ? PLoS One 11:e0160852. https://doi.org/10.1371/journal.pone.0160852
Khalaf EM, Raizada MN (2016) Taxonomic and functional diversity of cultured seed associated microbes of the cucurbit family. BMC Microbiol 16:131. https://doi.org/10.1186/s12866-016-0743-2
Kim H, Lee Y-H (2020) The Rice microbiome: a model platform for crop Holobiome. Phytobiomes J 4:5–18. https://doi.org/10.1094/PBIOMES-07-19-0035-RVW
Klaedtke S, Jacques M-A, Raggi L, Préveaux A, Bonneau S, Negri V, Chable V, Barret M (2016) Terroir is a key driver of seed-associated microbial assemblages: Terroir shapes the seed microbiota. Environ Microbiol 18:1792–1804. https://doi.org/10.1111/1462-2920.12977
Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6:1378–1390. https://doi.org/10.1038/ismej.2011.192
Kristin A, Miranda H (2013) The root microbiota—a fingerprint in the soil? Plant Soil 370:671–686. https://doi.org/10.1007/s11104-013-1647-7
Kumar J, Singh D, Ghosh P, Kumar A (2017) Endophytic and epiphytic modes of microbial interactions and benefits. In: Singh DP, Singh HB, Prabha R (eds) In plant-microbe interactions in agro-ecological perspectives. Springer Singapore, Singapore, pp 227–253
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. https://doi.org/10.1128/AEM.00335-09
Lejal E, Estrada-Peña A, Marsot M, Cosson JF, Rué O, Mariadassou M, Midoux C, Vayssier-Taussat M, Pollet T (2020) Taxon appearance from extraction and amplification steps demonstrates the value of multiple controls in tick microbiota analysis. Front Microbiol 11:1093. https://doi.org/10.3389/fmicb.2020.01093
Links MG, Demeke T, Gräfenhan T, Hill JE, Hemmingsen SM, Dumonceaux TJ (2014) Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol 202:542–553. https://doi.org/10.1111/nph.12693
Liu Y, Zuo S, Xu L, Zou Y, Song W (2012) Study on diversity of endophytic bacterial communities in seeds of hybrid maize and their parental lines. Arch Microbiol 194:1001–1012. https://doi.org/10.1007/s00203-012-0836-8
Love M, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Magan N, Aldred D, Mylona K, Lambert RJW (2010) Limiting mycotoxins in stored wheat. Food Additives Contaminants: Part A 27:644–650. https://doi.org/10.1080/19440040903514523
Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M (2014) Swarm: robust and fast clustering method for amplicon-based studies. PeerJ 2:e593. https://doi.org/10.7717/peerj.593
Mano H, Tanaka F, Watanabe A, Kaga H, Okunishi S, Morisaki H (2006) Culturable surface and endophytic bacterial flora of the maturing seeds of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ 21:86–100. https://doi.org/10.1264/jsme2.21.86
Matsumoto H, Fan X, Wang Y, Kusstatscher P, Duan J, Wu S, Chen S, Qiao K, Wang Y, Ma B, Zhu G, Hashidoko Y, Berg G, Cernava T, Wang M (2021) Bacterial seed endophyte shapes disease resistance in rice. Nature plants 7:60–72. https://doi.org/10.1038/s41477-020-00826-5
Mavrodi DV, Blankenfeldt W, Thomashow LS (2006) Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol 44:417–445. https://doi.org/10.1146/annurev.phyto.44.013106.145710
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Mendes R, Kruijt M, De Bruijn I et al (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100. https://doi.org/10.1126/science.1203980
Monnet C, Correia K, Sarthou A-S, Irlinger F (2006) Quantitative detection of Corynebacterium casei in cheese by real-time PCR. Appl Environ Microbiol 72:6972–6979. https://doi.org/10.1128/AEM.01303-06
Monnet C, Straub C, Castellote J, Onesime D, Bonnarme P, Irlinger F (2013) Quantification of yeast and bacterial gene transcripts in retail cheeses by reverse transcription-quantitative PCR. Appl Environ Microbiol 79:469–477. https://doi.org/10.1128/AEM.02360-12
Nabti E, Bensidhoum L, Tabli N, Dahel D, Weiss A, Rothballer M, Schmid M, Hartmann A (2014) Growth stimulation of barley and biocontrol effect on plant pathogenic fungi by a Cellulosimicrobium sp. strain isolated from salt-affected rhizosphere soil in northwestern Algeria. Eur J Soil Biol 61:20–26. https://doi.org/10.1016/j.ejsobi.2013.12.008
Nelson EB (2004) Microbial dynamics and interactions in the spermosphere. Annu Rev Phytopathol 42:271–309. https://doi.org/10.1146/annurev.phyto.42.121603.131041
Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356. https://doi.org/10.1128/MMBR.00051-12
Ofek M, Hadar Y, Minz D (2011) Colonization of cucumber seeds by bacteria during germination: bacterial colonization of cucumber seeds. Environ Microbiol 13:2794–2807. https://doi.org/10.1111/j.1462-2920.2011.02551.x
ONU (2019) Deux milliards de personnes de plus sur la terre en 2050. In: Nations Unies. Département des affaires économiques et sociales. https://www.un.org/development/desa/fr/news/population/world-population-prospects-2019.html. Accessed 12 Oct 2020
Pancher M, Ceol M, Corneo PE, Longa CMO, Yousaf S, Pertot I, Campisano A (2012) Fungal endophytic communities in grapevines (Vitis vinifera L.) respond to crop management. Appl Environ Microbiol 78:4308–4317. https://doi.org/10.1128/AEM.07655-11
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci 110:6548–6553. https://doi.org/10.1073/pnas.1302837110
Plotnikov AO, Balkin AS, Gogoleva NE, Lanzoni O, Khlopko YA, Cherkasov SV, Potekhin AA (2019) High-throughput sequencing of the 16S rRNA gene as a survey to analyze the microbiomes of free-living ciliates Paramecium. Microb Ecol 78:286–298. https://doi.org/10.1007/s00248-019-01321-x
Rascovan N, Carbonetto B, Perrig D, Díaz M, Canciani W, Abalo M, Alloati J, González-Anta G, Vazquez MP (2016) Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci Rep 6:28084. https://doi.org/10.1038/srep28084
Rezki S, Campion C, Iacomi-Vasilescu B, Preveaux A, Toualbia Y, Bonneau S, Briand M, Laurent E, Hunault G, Simoneau P, Jacques MA, Barret M (2016) Differences in stability of seed-associated microbial assemblages in response to invasion by phytopathogenic microorganisms. PeerJ 4:e1923. https://doi.org/10.7717/peerj.1923
Ritpitakphong U, Falquet L, Vimoltust A et al (2016) The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 210:1033–1043. https://doi.org/10.1111/nph.13808
Rochefort A, Simonin M, Marais C, et al (2020) Asymmetric outcome of community coalescence of seed and soil microbiota during early seedling growth. bioRxiv https://doi.org/10.1101/2020.11.19.390344
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Rosenblueth M, López-López A, Martínez J et al (2012) Seed bacterial endophytes: common genera, seed-to-seed variability and their possible role in plants. Intern. Symposium:39–48. https://doi.org/10.17660/ActaHortic.2012.938.4
Rybakova D, Mancinelli R, Wikström M, Birch-Jensen AS, Postma J, Ehlers RU, Goertz S, Berg G (2017) The structure of the Brassica napus seed microbiome is cultivar-dependent and affects the interactions of symbionts and pathogens. Microbiome 5:104. https://doi.org/10.1186/s40168-017-0310-6
Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. https://doi.org/10.1186/s12915-014-0087-z
Schneider A, Flénet F, Dumans P, Bonnin E, de Chezelles E, Jeuffroy MH, Hayer F, Nemecek T, Carrouée B (2010) Diversifier les rotations céréalières notamment avec du pois et du colza – Données récentes d’expérimentations et d’études. OCL 17:301–311. https://doi.org/10.1051/ocl.2010.0332
Schouten A, Van den Berg G, Edel-Hermann V et al (2004) Defense responses of Fusarium oxysporum to 2, 4-diacetylphloroglucinol, a broad-spectrum antibiotic produced by Pseudomonas fluorescens. Mol. Plant microbe Interact. 17:1201–1211. https://doi.org/10.1094/MPMI.2004.17.11.1201
Shade A, Jacques M-A, Barret M (2017) Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr Opin Microbiol 37:15–22. https://doi.org/10.1016/j.mib.2017.03.010
Shata HMA (2018) Statistical optimization of erythromycin production by Saccharopolyspora erythraea under solid state fermentation of agro-industrial materials using response surface methodology. J. Microbiol. Biotechnol. Food Sci. 8:692–697. https://doi.org/10.15414/jmbfs.2018.8.1.692-697
Shimokawatoko Y, Sato N, Yamaguchi T, Tanaka H (2012) Development of the novel insecticide Spinetoram (DIANA®). 14
Smits THM, Rezzonico F, Kamber T, Goesmann A, Ishimaru CA, Stockwell VO, Frey J̈E, Duffy B (2010) Genome sequence of the biocontrol agent Pantoea vagans strain C9-1. J Bacteriol 192:6486–6487. https://doi.org/10.1128/JB.01122-10
Sultanpuram VR, Mothe T, Chintalapati S, Chintalapati VR (2015) Cellulosimicrobium aquatile sp. nov., isolated from Panagal reservoir, Nalgonda, India. Antonie Van Leeuwenhoek 108:1357–1364. https://doi.org/10.1007/s10482-015-0588-y
Thiebeau P, Lô-Pelzer E, Klumpp K et al (2010) Conduite des légumineuses pour améliorer l’efficience énergétique et réduire les émissions de gaz à effet de serre à l’échelle de la culture et de l’exploitation agricole. Innovations Agronomiques 11:45–58
Torres-Cortés G, Garcia BJ, Compant S, Rezki S, Jones P, Préveaux A, Briand M, Roulet A, Bouchez O, Jacobson D, Barret M (2019) Differences in resource use lead to coexistence of seed-transmitted microbial populations. Sci Rep 9:6648. https://doi.org/10.1038/s41598-019-42865-9
Truyens S, Weyens N, Cuypers A, Vangronsveld J (2015) Bacterial seed endophytes: genera, vertical transmission and interaction with plants: bacterial seed endophytes. Environ Microbiol Rep 7:40–50. https://doi.org/10.1111/1758-2229.12181
Tsuge T, Harimoto Y, Akimitsu K, Ohtani K, Kodama M, Akagi Y, Egusa M, Yamamoto M, Otani H (2013) Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol Rev 37:44–66. https://doi.org/10.1111/j.1574-6976.2012.00350.x
Vandenkoornhuyse P, Quaiser A, Duhamel M, le van A, Dufresne A (2015) The importance of the microbiome of the plant holobiont. New Phytol 206:1196–1206. https://doi.org/10.1111/nph.13312
Verce M, De Vuyst L, Weckx S (2019) Shotgun metagenomics of a water kefir fermentation ecosystem reveals a novel Oenococcus species. Front Microbiol 10:479. https://doi.org/10.3389/fmicb.2019.00479
Verma AK, Agrawal K (2017) In vitro evaluation of antibacterial activity of some medicinal plants against Xanthomonas pisi causing leaf spot of pea. Int J Pharm Sci Rev Res 45:156–159
Wassermann B, Cernava T, Müller H, Berg C, Berg G (2019) Seeds of native alpine plants host unique microbial communities embedded in cross-kingdom networks. Microbiome 7:108. https://doi.org/10.1186/s40168-019-0723-5
Weissenbach J, Sghir A (2016) Microbiotes et métagénomique. Med Sci (Paris) 32:937–943. https://doi.org/10.1051/medsci/20163211008
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer. https://doi.org/10.18637/jss.v077.b02
Yang L, Danzberger J, Schöler A, Schröder P, Schloter M, Radl V (2017) Dominant groups of potentially active bacteria shared by barley seeds become less abundant in root associated microbiome. Front Plant Sci 8:1005. https://doi.org/10.3389/fpls.2017.01005
Yoon JH, Kang SJ, Schumann P, Oh TK (2007) Cellulosimicrobium terreum sp. nov., isolated from soil. Int J Syst Evol Microbiol 57:2493–2497. https://doi.org/10.1099/ijs.0.64889-0
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Zhang Z, Nan Z (2014) Erwinia persicina, a possible new necrosis and wilt threat to forage or grain legumes production. Eur J Plant Pathol 139:349–358. https://doi.org/10.1007/s10658-014-0390-0
Acknowledgements
We thank the INRAE MIGALE bioinformatics platform (https://migale.inrae.fr/) for providing computational resources and support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Editorial Responsibility: Luz E. de-Bashan
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
Figure S1 Clustering of 49 bacterial strains isolated from pea samples by metabolic properties based on API 50 CH and API ZYM. Dark black indicates carbon sources and enzyme activities with a positive reaction in the API50 and APIZYM panels; white indicates a negative reaction. The bacterial strains were clustered by an Ascending Hierarchical Classification and color-coded by cluster. The 15 species detected by metabarcoding analysis were noticed in red. Fig. S2 Total viable count of microorganisms in pea seeds from six cooperatives (A, B, C, D, E and F) located in three different countries (France, Sweden and Canada), over five days of soaking on different isolation media. The MRS (De Man, Rogosa and Sharpe), BHI (Brain Heart Infusion) and YEGC (Yeast Extract Glucose Chloramphenicol) agar media are suitable for lactic acid bacteria, mesophilic aerobic bacteria (medium BHI) and fungi (molds and yeasts), respectively. Only the averages of triplicates are given with the standard errors of the mean (SEM) (PDF 144 kb)
ESM 2
Figure S3 Rarefaction curves of V3/V4 Miseq sequencing reads of the bacterial 16S rRNA gene from 16 samples (a) and the ribosomal internal transcribed spacer 2 (ITS2) region for fungi from 17 samples (b) of soaked pea seeds from different cooperatives (A, B, C, D, E and F) located in three different countries (France, Canada and Sweden) (PDF 48 kb)
ESM 3
Table S1 The specific meteorological data (temperature, precipitation, humidity, sunshine and winds) of the countries (France, Sweden and Canada) of cultivation and cooperatives (A, B, C, D, E and F) in 2017. Data were taken from the NASA website (https://power.larc.nasa.gov/data-access-viewer/) (XLSX 10 kb)
ESM 4
Table S2 List of 101 microorganisms isolated from pea seeds from six cooperatives (A, B, C, D, E and F) located in three different countries (France, Canada and Sweden). Origin (country and cooperative) and culture conditions (agar medium, temperature and anaerobic and aerobic conditions) are indicated. The identification of bacteria was determined by comparison of the 16S rRNA gene sequences to the EZ Biocloud database, and the identification of the yeasts was determined by comparison of the 26S rRNA gene sequences (D1D2 region) to the Genbank database using the Basic Local Alignment Search Tool (BLAST) (XLSX 23 kb)
ESM 5
Table S3 Metabolic properties of the three fungal strains isolated from pea seeds based on the API Candida and API ZYM galleries. Dark black indicates carbon sources and enzyme activities with a positive reaction in the API Candida and API ZYM panels; white indicates a negative reaction (XLSX 11 kb)
ESM 6
Table S4 Summary of information regarding the runs performed with the MiSeq sequencing platform and bio-informatics analysis. The number of paired-reads, quality sequences, operational taxonomic units (OTUs), abundant OTUs (aOTUs representing at least 0.005% of the library size) and abundant quality sequences are indicated for each seed sample and in each column. In the column, % of sequences belonging to contaminating OTUs, a represents the sequences occurring in contaminating OTUs other than Cellulosimicrobium, and b represents the sequences belonging to the OTU of Cellulosimicrobium. * Samples with less than 1000 sequences were eliminated from the analysis. (XLSX 14 kb)
ESM 7
Table S5 Differentially abundant microbial OTUs between Canadian and French peas. The 7 bacterial and 13 fungal species were represented by more than 0.5% of all reads. The log2FoldChange is the difference in cluster detection between the two origins (Canada to France). When it is positive, the cluster is more abundant in French peas than in Canadian peas, and the higher it is, the higher the difference in detection is. When negative, the cluster is more abundant in Canadian peas than in French peas. Padj = p value adjusted by the Benjamini-Hochberg method proposed within the DESeq2 package. Only OTUs with a padj <0.05 were considered as differentially abundant between sample groups. * Species belonging to core microbiome. (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Chartrel, V., Dugat-Bony, E., Sarthou, AS. et al. The microbial community associated with pea seeds (Pisum sativum) of different geographical origins. Plant Soil 462, 405–427 (2021). https://doi.org/10.1007/s11104-021-04856-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-021-04856-6