Abstract
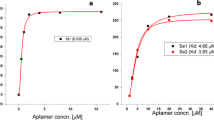
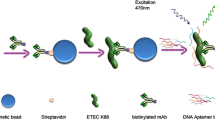
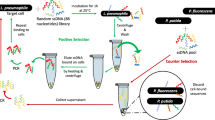
The present study demonstrates, development of ssDNA aptamers against whole cell of S. flexneri employing a whole bacterium-based Systemic Evolution of Ligands by Exponential Enrichment (SELEX). After ten rounds of SELEX, cell surface specific aptamer pool was cloned, sequenced and divided based on sequence similarities and secondary structure. Binding affinity of FITC labelled aptamer from different group were carried out by flow cytometry analysis. The dissociation constant (Kd) values for specific and higher binder were evaluated to range from 144 to 329 nM. Six high binding aptamers with lower dissociation constant was chosen for selectivity study. Aptamer SHI 23, SHI 37 and SHI 42 showed higher selectivity towards S. flexneri in comparison with other related bacteria. Further applicability of selected aptamer was proven by fluorescence assay for convenience detection of target cell from spiked water sample and natural contaminated water samples. Altogether, aptamer generated in this study can be alternative DNA ligands for detection of S. flexneri compared to available ligands.









Similar content being viewed by others
Data Availablity
Not applicable.
References
Niyogi SK (2005) Shigellosis. Journal of microbiology (Seoul, Korea) 43.2: 133–143
Bardhan P, Faruque ASG, Naheed A, Sack DA (2010) Decreasing shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerg Infect Dis 16, no. 11: 1718-1723
Sur D, Ramamurthy T, Deen J, Bhattacharya SK (2004) Shigellosis: challenges & management issues. Ind J Med Res 120(5):454
Bennish ML (1991) Potentially lethal complications of shigellosis. Rev Infectious Diseas 13, no. Supplement 4: S319-S324
Bennish ML, Salam MA, Hossain MA, Myaux J, Khan EH, Chakraborty J, Henry F, Ronsmans C (1992) Antimicrobial resistance of Shigella isolates in Bangladesh, 1983–1990: Increasing frequency of strains multiply resistant to ampicillin, trimethoprim-sulfamethoxazole, and nalidixic acid. Clinical infectious diseases 14, no 5: 1055–1060
Akiba T, Koyama K, YoshitoIshiki SK, Fukushima T (1960) On the mechanism of the development of multiple-drug-resistant clones of shigella. Japanese j microbiol 4(2):219–227
McMahan L, Grunden AM, Devine AA, Sobsey MD (2012) Evaluation of a quantitative H 2 S MPN test for fecal microbes analysis of water using biochemical and molecular identification. Water res 46, 1693(6):–1704
Mokhtari W, Nsaibia S, Gharbi A, Aouni M (2013) Real-time PCR using SYBR green for the detection of Shigella spp. in food and stool samples. Mol Cell Probes 27(1):53–59
Robbins JB, Kubler-Kielb J, Vinogradov E, Mocca C, Pozsgay V, Shiloach J, Schneerson R (2009) Synthesis, characterization, and immunogenicity in mice of Shigella sonnei O-specific oligosaccharide-core-protein conjugates. Proc National Acad Sci 106(19):7974–7978
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249(4968):505–510
Centi S, Messina G, Tombelli S, Palchetti I, Mascini M (2008) Different approaches for the detection of thrombin by an electrochemical aptamer-based assay coupled to magnetic beads. Biosens Bioelectron 23(11):1602–1609
Jayasena SD (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clinic chem 45(9):1628–1650
Strehlitz B, Reinemann C, Linkorn S, Stoltenburg R (2012) Aptamers for pharmaceuticals and their application in environmental analytics. Bioanal Rev 4(1):1–30
Radom, Filip, Przemysław M. Jurek, Maciej P. Mazurek, JacekOtlewski, and Filip Jeleń. "Aptamers: molecules of great potential." Biotechnol Adv 31, no. 8 (2013): 1260–1274
Meyer M, Scheper T, Walter J-G (2013) Aptamers: versatile probes for flow cytometry. Appl microbiol biotechnol 97(16):7097–7109
Setlem K, Mondal B, Ramlal S, Kingston J (2016) Immuno affinity SELEX for simple, rapid and cost-effective Aptamer enrichment and identification against Aflatoxin B1. Front Microbiol 7:1909
Mondal B, Ramlal S, Lavu PSR, Murali HS, Batra HV (2015) A combinatorial systematic evolution of ligands by exponential enrichment method for selection of aptamer against protein targets. Appl microbiol biotechnol 99(22):9791–9803
Hamula CLA, Zhang H, Li F, Wang Z, Le XC, Li X-F (2011) Selection and analytical applications of aptamers binding microbial pathogens. TrAC Trends Anal Chem 30(10):1587–1597
Jenison RD, Gill SC, Pardi A, Polisky B (1994) High-resolution molecular discrimination by RNA. Sci-Newyork Washington 263:1425–1425
Meyer S, Maufort JP, Nie J, Stewart R, McIntosh BE, Conti LR, Ahmad KM, Soh HT, Thomson JA (2013) Development of an efficient targeted cell SELEX procedure for DNA aptamer reagents. PLoS One 8(8):e71798
Cibiel A, MiottoDupont D, Ducongé F (2011) Methods to identify aptamers against cell surface biomarkers. Pharmaceuticals 4(9):1216–1235
Moon J, Kim G, Park S (2014) Development of ssDNA aptamers for the capture and detection of Salmonella typhimurium. Anal Methods 6(18):7442–7448
Duan N, Wu S, Chen X, Huang Y, Wang Z (2012) Selection and identification of a DNA aptamer targeted to Vibrio parahemolyticus. J Agric Food Chem 60(16):4034–4038
Lavu PSR, Mondal B, Ramlal S, Murali HS, Batra HV (2016) Selection and Characterization of Aptamers Using a Modified Whole Cell Bacterium SELEX for the Detection of Salmonella enterica Serovar Typhimurium. ACS combinatorial science
Gong W, Duan N, Wu S, Huang Y, Chen X, Wang Z (2015) Selection, identification, and application of dual DNA aptamers against Shigella sonnei. Anal Methods 7(8):3625–3631
Cao X, Li S, Chen L, Ding H, HuaXu YH, Li J et al (2009) Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res 37(14):4621–4628
Kim YS, Song MY, Jurng J, Kim BC (2013) Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell–systematic evolution of ligands by exponential enrichment approach. Anal Biochem 436(1):22–28
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Zuker M (1989) On finding all suboptimal folding of an RNA molecule. Science 244(4900):48–52
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415
Meyer C, Hahn U, Rentmeister A (2011) Cell-specific aptamers as emerging therapeutics. Journal of nucleic acids 2011:1–18
Shamah SM, Healy JM, Cload ST (2008) Complex target SELEX. Acc Chem Res 41(1):130–138
Cerchia L, Ducongé F, Pestourie C, Boulay J, Aissouni Y, Gombert K, Tavitian B, De Franciscis V, Libri D (2005) Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoSBiol 3(4):e123
Pestourie C, Cerchia L, Gombert K, Aissouni Y, Boulay J, De Franciscis V, Libri D, Tavitian B, Ducongé F (2006) Comparison of different strategies to select aptamers against a transmembrane protein target. Oligonucleotides 16(4):323–335
Berezovski MV, Musheev MU, Drabovich AP, Jitkova JV, Krylov SN (2006) Non-SELEX: selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat Protoc 1(3):1359–1369
Acknowledgments
The first author is indebted to Department of Science and Technology (DST, New Delhi, India), for financial assistance through women scientist program. The second author thankful to Department of Science and Technology (DST, New Delhi, India), for financial assistance through Inspire programme. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The corresponding author and first author design, data collection and analysis, decision to publish, or preparation of the manuscript.
Code Availability
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Dr. Padma Sudharani Lavu was involved in research designing, performing experiment, and manuscript writing. Bhairab Mondal was involved in research designing, performing experiment, data analysis, and manuscript writing. Dr. Shylaja Ramlal was involved in research designing, manuscript writing and correction of manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Informed Consent
Not applicable.
Conflict of Interest
All authors declare no conflicts of interest.
Supplementary Information
ESM 1
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Lavu, P.S., Mondal, B. & Ramlal, S. Selection and Characterization of Cell Surface Specific Aptamer and Development of Fluorescence Assay for Detection of Shigella flexneri from Water Samples. J Fluoresc 31, 685–693 (2021). https://doi.org/10.1007/s10895-021-02691-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-021-02691-7