Abstract
Knowledge of the degree of genetic diversity can provide fundamental information to breeders for use in various breeding programmes, for instance for the selection of lines with better adaptability. The genetic diversity analysis of 188 winter wheat genotypes demonstrated that this group of cultivars could be divided into four clusters based primarily on geographical origin. The first group contained mostly American and Asian cultivars, while cluster 2 consisted of Central European cultivars, cluster 3 of Hungarian and South European cultivars and cluster 4 mainly of cultivars from Western Europe. Cultivars used in breeding programmes in Central and South East European breeding programmes were found in all four clusters. Wheat genotypes originating from this region of Europe proved to have greater genetic variability than lines from Western and Northern Europe. Among the four clusters, there were also differencies in the frequencies of winter–spring alleles in Vrn-A1, Vrn-B1, Vrn-D1 vernalisation response genes and in the frequencies of sensitive–insensitive alleles in Ppd-B1 and Ppd-D1 photoperiod response genes, which explained the differences in heading date of the four clusters as well.
Similar content being viewed by others
Introduction
Breeding programmes will only be successful if the cultivars have sufficient genetic diversity, since this improves the chance that promising traits will be manifested, thus ensuring the selection of lines with better adaptability (Bouffier et al. 2008; Karsai et al. 2012; Orabi et al. 2014). Theoretically, intensive targeted selection may reduce the number of rare alleles, with a consequent decline in genetic diversity (Tanksley and McCouch 1997; Fu et al. 2006; Lopes et al. 2015). This may result in the loss of numerous traits that could be used to improve agronomic and quality characters or to breed genotypes better adapted to various environmental stress factors, such as resistance to pathogens or tolerance of the extreme conditions caused by climate change (Smale 1997; Chao et al. 2008; Charmet 2011; Rauf et al. 2010; Tester and Langridge 2010; van de Wouw et al. 2010; Benson et al. 2012; Lopes et al. 2015). Knowledge of the genetic diversity existing between various breeding lines could therefore provide important information for breeders. Roussel et al. (2005) demonstrated an increase in the degree of genetic similarity between European wheat cultivars, but a difference could be observed between the geographical regions of Europe as regards the allele distribution patterns, with a lower number of allele variations in Western Europe than in Central and South Eastern Europe (Roussel et al. 2005; Balfourier et al. 2007; Hai et al. 2007; Zhang et al. 2011; Kiss et al. 2014; Novoselović et al. 2016; El-Esawi et al. 2018). This was confirmed by allele diversity analyses performed using SSR markers, in which a lower allele number was detected in Western European countries (France, the Netherlands, Great Britain, Belgium) compared with South Eastern Europe (countries previously part of Yugoslavia, Greece, Bulgaria, Romania, Hungary) and the Mediterranean regions (Italy, Spain, Portugal) (Roussel et al. 2005; Balfourier et al. 2007). This can be explained by the diverse environmental conditions and soil factors and by the differences in breeding practices (Stachel et al. 2000; Huang et al. 2002; Roussel et al. 2005; El-Esawi et al. 2018). Nevertheless, many authors have published contradictory results. Huang et al. (2007) used 42 microsatellite markers to analyse 511 Central and Northern European cultivars widely cultivated between 1945 and 2000 and found no significant quantitative decrease in genetic diversity. Similar results were reported by White et al. (2008) in a study on the genetic diversity of British, American and Australian cultivars. Due to the increasing importance of this topic, future papers will hopefully help to clarify the contradictions currently found in the international literature.
Genetic diversity can be characterised either by indirectly estimating genetic distances or determining morphological and phenotypic traits, or by a direct comparison of differences in the DNA sequences of the genotypes using molecular markers (Astarini et al. 2004; Fufa et al. 2005; Zhang et al. 2011; Spanic et al. 2016). Indirectly acquired information is not always sufficiently reliable for genetic characterisation due to deficiencies in the pedigree data, the disregard of natural and artificial selection or of mutations, and the environment-dependent variability of phenotypic traits (Parker et al. 2002; Almanza-Pinzón et al. 2003; Reif et al. 2005). By contrast, molecular marker systems are reliable, as they are not influenced by environmental, pleiotropic or epistatic effects, and DNA extracts isolated from any type of tissue in any phenophase can be used for the analysis (Mukhtar et al. 2002; Fufa et al. 2005; Karsai et al. 2012; El-basyoni et al. 2013). Over the last decade, there has been a great increase in the number of genetic diversity studies performed using a reliable marker system (RFLP, AFLP, RAPD, SSR, SNP or DArT) on various plant species (Siedler et al. 1994; Röder et al. 2002; Khan et al. 2005; Stodart et al. 2005; Roussel et al. 2005; White et al. 2008; Benson et al. 2012; Matthies et al. 2012; Nielsen et al. 2014; Kabbaj et al. 2017; El-Esawi et al. 2018). The Diversity Arrays Technology (DArT) is a rapid, cost-effective high-throughput marker system for whole genome analysis, which does not require preliminary knowledge of the sequences. This has made it one of the most widely used marker technologies for genetic analysis (Jaccoud et al. 2001; Zhang et al. 2011; Ficco et al. 2012; El-Esawi et al. 2018). The first wheat genome association map prepared with this method was published by Crossa et al. (2007). The DArT marker system has now been applied on numerous species, including cereals such as barley (Hordeum vulgare L.), wheat (Triticum aestivum L.) and durum wheat (Triticum durum L.) (Zhang et al. 2011). This technology can be used not only to prepare high-resolution genetic maps and perform association analyses, but can also help scientists to obtain a better understanding of the extent of genetic diversity and of population genetics correlations (Semagn et al. 2006; Crossa et al. 2007; White et al. 2008; Raman et al. 2010; Zhang et al. 2011).
The distribution pattern of allele variants of the major genes responsible for the environmental adaptation of wheat (the VRN: vernalisation requirement and PPD: photoperiod sensitivity genes responsible for the vegetative–generative transition) could be especially important for the toleration of climatic extremes, because the length and timing of the vegetative–generative transition can be regulated by altering the allele combination of these genes, and this could be of decisive importance in the achievement of satisfactory yield potential (González et al. 2005; Borràs et al. 2009; Chen et al. 2009, 2010; Kiss et al. 2014). One of the most important steps in the adaptation process is the heading date, which is determined to a significant extent by these genes (Dubcovsky et al. 1998; Worland, 1996). In the case of wheat, several gene families are involved in the genetic control of the vernalisation requirement, among which the Vrn-A1, Vrn-B1 and Vrn-D1 genes have the greatest effect (Pugsley, 1971, 1972). Depending on the ratio of dominant and recessive alleles of the Vrn genes in the three genomes of hexaploid wheat, a distinction can be made between winter (recessive) and spring (dominant) cultivars, while genotypes with a facultative growth habit have various combinations of dominant and recessive alleles. The influence of the dominant alleles of these genes in spring forms means that the plants require little or no cold treatment for the transition to the generative stage (Pugsley 1971, 1972; Kato et al. 2001; Loukoianov et al. 2005). In wheat, the most influential genes in the regulation of photoperiod sensitivity are Ppd-D1 and Ppd-B1 (Law et al. 1978; Börner et al. 1993). The dominant photoperiod-insensitive allele type results in early flowering under short-day conditions, while the heading of genotypes carrying the photoperiod-sensitive allele type is retarded or may not take place at all under such conditions (Worland 1996). In the case of the Vrn-A1, Vrn-B1, Vrn-D1 and Ppd-D1 genes, length polymorphism in the promoter, intron and exon regions forms the genetic background of the spring or photoperiod-insensitive allele types (Fu et al. 2005; Beales et al. 2007), while the photoperiod-insensitive allele of the Ppd-B1 can be attributed to the enhanced quantity of gene products due to the presence of extra gene copies (Díaz et al. 2012). In addition, it was identified that the intercopy structure between the duplicated genes of Ppd-B1 also significantly determines the level of photoperiod sensitivity, the effect of which can be detected in heading dates under field conditions especially in association with the CNV (Kiss et al. 2014). Under field conditions, however, the phenotypic effects of the various alleles of these genes exhibit considerable variability resulting from the differing environmental effects experienced in different years, leading to contradictory results and to a dearth of information on the frequency distribution of the allele combinations of the three VRN1 and the two PPD1 genes (Snape et al. 1985; Worland 1996; Blake et al. 2009; Andeden et al. 2011; Díaz et al. 2012).
The main aim of the present work was to determine (1) the degree of genetic diversity and (2) the population structure of wheat cultivars of diverse origin, and (3) the allele distribution of each subgroup of the major genes responsible for the environmental adaptability of wheat (VRN and PPD). A high-resolution marker technology (DArT), the determination of copy number (for the PPD-B1 gene) and allele-specific markers for the major plant development genes (VRN-A1, VRN-B1, VRN-D1, PPD-B1, PPD-D1) were used in the study. It is hoped that the resulting knowledge on genetic diversity and population structure will be of use in future breeding programmes.
Materials and method
The 188 wheat genotypes included in the experiment were obtained from the cereal gene bank of the ATK Agricultural Institute and were chosen on the basis of previous flowering data (Supplemental Table 1). The samples include both old cultivars that were previously widely cultivated and newly bred genotypes that are of importance for today’s wheat production. The field heading date (Z59: spike out of the flag leaf sheath) of the genotypes was observed in three consecutive years (2013–2015) in Martonvásár on chernozem soil with average N, P2O5 and K2O contents, based on Tottman and Makepeace (1979). The heading date was determined as the days from January 1st, corrected with the mean vernalisation requirement and photoperiod (effective heat sum) of the plants, using the method of Bogard et al. (2015). The effective heat sum is the sum of the mean daily temperatures after the saturation of the vernalisation requirement, modified by the day length, i.e. the total heat quantity obtained by the plant up to the given phenophase (SPTV = TT × FV × FP, where TT is the total daily heat sum, FV the vernalisation factor and FP the photoperiod factor).
The fresh shoot samples (100 mg) used for the extraction of genomic DNA were digested in liquid nitrogen using buffers from the DNeasy® Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. A list of the gene-specific primers is given in Appendix S2, together with the relevant references, which were used to evaluate the allele compositions of each wheat genotype in the vernalisation (Vrn1 genes) and photoperiod response genes (Ppd1 genes). In the case of the PPD-B1 gene, the copy number variation (CNV) was determined with the Multiplex TaqMan® Assay at iDna Genetics Ltd. (Norwich Research Park, Norwich, UK) (Díaz et al. 2012), in addition to checking for the presence of the Chinese Spring type intercopy structure, which also influences the level of photoperid sensitivity in this gene (Kiss et al. 2014). The DArT analysis of the DNA samples was performed by the Triticarte subsidiary of Diversity Arrays Technology Pty Ltd. (CSIRO, 1 Wilf Crane Crescent, Yarralumla, ACT 2600, Australia) (http://www.triticarte.co.au). The polymorphism information content (PIC) values of the individual DArT markers were calculated with the help of the formula: PIC = 1 – Σ(Pi)2, where Pi is the frequency of the given allele or locus (Anderson et al. 1993). A binary matrix was then created from the DArT data, where the presence or absence of the specific marker alleles was designated as 1 or 0. A difference matrix was then compiled from the binary data using Jaccard’s distance coefficient [JDC = 1 – a/(n–d), where a is the number of marker fragments common to the two genotypes, n is the total number of marker fragments and d is the number of marker fragments missing from both genotypes]. The comparison of individual pairs was performed using a 0–1 scale, where 0 represented complete identity and 1 complete difference. The relationship matrix was determined with the UPGMA (unweighted pair group method using the arithmetic mean) module of TASSEL 3.0 (trait analysis by association, evolution and linkage), and this was used to compile dendrograms. The population structure was determined with the help of the STRUCTURE 2.3.4 program (Pritchard et al. 2000) on the data matrix of 184 genotypes × 249 markes. For predicting the most probable number of subgroups within the population, the probabilities (K) were tested for the presence of subgroups from 1 to 10 in 10 independent runs at each step using a burn in period of 100,000. The most probable subgroup number was identified at 4, where ΔK was the highest. Using the Q values (ranging from 0 to 1), which indicate the probability with which a genotype can be assigned to a particular subgroup (Q matrix), all the cultivars were assigned to one of the 4 subgroups. Principal component analysis (PCA) was performed with the help of the Statistica 6 software package (StatSoft Inc., Tulsa, OK, USA) on the data matrix of the mean values of the three-year heading data (Z59 developmental phase) and the Q values of the studied genotypes. In order to examine whether the geographic origin has any role in formation of the population structure, cultivars were grouped into six large geographic regions based on their origin. These were European, Asian, American, Australian and African. Rank correlation was then carried out between the group numbers of geographic regions and the Q values of population structure.
Results
Genetic diversity and population structure of the hexaploid wheat panel
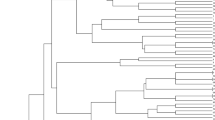
The analysis of the population structure and genetic diversity of the association panel was based on DArT markers. Among the 4606 DArT markers, 1642 proved to be polymorph for the 188 wheat cultivars, of which the chromosomal localisation of 970 is known. To examine the population structure of the tested cultivars, 249 of these markers were selected; these were independent of each other, exhibited well-balanced segregation frequency and were evenly spaced over the genome. The genotypes could be divided into four clusters (Figs. 1, 2) using the Q matrix method in the STRUCTURE program. The Q values, which range from 0 to 1, indicate the probability with which a genotype can be assigned to a particular cluster. The higher this number, the more reliable the genotype assignment. All 970 of the DArT markers with known chromosomal localisation were used for the genetic diversity analysis. Five of the wheat cultivars did not give a measurable response, so these were omitted from further analysis.
Relationships and population structure of the wheat genotypes examined
59:SAROZ; 117:KLEIN-CASTOR; 76:INIA-TORCAZA; 116:KLEIN-CAPRICORNIO; 191:APS1P-ADE; 119:KLEIN-ESCUDO; 104:BUCK-75ANIVERSARIO; 74:BUCK-PANADERO; 105:BUCK-SURENO; 101:BLASCO; 37:MV-HOMBAR; 113:FLEMING; 51:MADSEN; 138:PBV2; 82:993-11-SGP1; 91:ALTAY-2000; 126:MAESTRA; 62:YILDIZ; 19:SULTAN95; 56:ESER; 11:SPADA; 55:BAYRAKTAR; 118:KLEIN-CHAJA; 102:BIGGAR; 38:LONA; 141:RED-RIVER-68; 58:MVSW33-05; 107:CHARA; 197:SUNSTAR; 134:NORDIC; 83:P306; 92:BAI-HUO; 135:NUO-MAIZI; 123:KUKRI; 32:GLENLEA; 152:WILDCAT; 103:BONCAP; 120:KLEINFLECHA; 131: ND495; 195:MARQUIS; 40:KATEPWA; 124:LAURA-CAN; 47:HALLAM; 88:NUDAKOTA; 64:FLEISCHMANN-481; 145:SIRBAN-PROLIFIK; 137:PAN2001-27; 13:AGENT; 49:JIAN155; 24: LANGFANG-3; 153:ZHONG-MAI-175; 20:VIATOR; 4:MVEMMA; 89:ABONY; 44:KINA9204; 172:UKRAINKA; 187:MV-KARIZMA; 28:YUMAI-10; 63:YUMAI-21; 168:SIMONIDA; 21:BRUTUS; 50:KG-KUNHALOM; 95:BALETKA; 90:ARIDA; 73:HP-PUSZTASZEL; 93:BALADA; 150:VLASTA; 65:BREA; 71:HUNOR; 79:MV27-07; 196:NOBEOKABOZU-KOMUGI; 173: MV-BERES; 161:LUDWIG; 193:MV213-10; 186:MV-TOLDI; 185:MV-TOBORZO; 189:MV-WALZER; 181:MV-MAZURKA; 178:MV-KODMON; 183:MV-SUVEGES; 182:MV-MENUETT; 175:MV-CSARDAS; 188:MV-VERBUNKOS; 80:RONA; 157:BRIANA; 128:MV15-06; 176:MV-KOKARDA; 26:MV17-09; 12:MV-VILMA; 129:MV-AMANDA; 14:MV-MARTINA; 87:MV23-09; 180:MV MARSALL; 159:DUMBRAVA; 162:MV-TALTOS; 130:MV-PRIZMA; 177:MV-KIKELET; 167:SANPASTORE; 122:KRASOTA; 200:GERONIMO; 3:MV-MAGMA; 36:MV-GORSIUM; 174:MV-BODRI; 86:PERVITSA; 6:MV-PALOTAS; 8:JUBILEJNAJA-50; 35:AURA; 125:KRASNODARSKAYA-99; 199:MV-LUCILLA; 169:SKOPJANKA; 61:TURKMEN; 109:DIVANA; 54:ARMCIM; 97:BEI-JING-0045; 127:MCNAIR-701; 27:ROANE; 184:MV-TALLER; 5:MV-SUMMA; 7:MV-MATYO; 114:GK-GONCOL; 17:GKHEJA; 46:GK-FENY; 84:GK-BERENY; 69:GK-CSILLAG; 68:GK-BEKES; 85:GK-HAJNAL; 156:BABUNA; 163:NS-RANA-1; 115:GOLUBICA; 143:RUZICA; 144:SAGITTARIO; 98:BILANCIA; 139:RAVENNA; 81:NW98S097; 41:SALAMOUNI; 160:LIBELLULA; 57:FENG-YOU-3; 190:YUMAI-34; 155:ADRIANA; 158:DEMETRA-OS; 154:ZLATNA-DOLINA; 164:SANA; 18:GK-SZALA; 42:COURTOT; 77:NOMADE; 170:SOISSONS; 121:KORELI; 179:MV-KOLOMPOS; 16:MV-VEKNI; 30:RENAN; 23:ALAMOOT; 29:ORH010918; 31:NZ4321-114; 147:SPARTACUS; 194:CADENZA; 192:KWS-SCIROCCO; 39:VANEK; 198:AMOR; 78:ORNICAR; 140:RECITAL; 9:DISPONENT; 10:ASPEN; 15:BURATINO; 43:LUPUS; 111:ESTEVAN; 108:CUTTER; 45:PANNONIKUS; 151:WENZEL; 75:HEREWARD; 94:BALANCE; 146:SOLSTICE; 149:VALORIS; 96:BASTIDE; 136:ORDEAL; 142:RIGI; 112:FERIA; 171:TOMMI; 48:CUBUS; 22:HAJDUSAG; 110:ELLVIS; 148:TIGER; 25:GK-HATTYU; 72:KG-BENDEGUZ
The first cluster contained 26 American, 9 European, 14 Asian, 3 Australian and 2 African cultivars, while there were 2 American, 52 European and 3 Asian genotypes in the second cluster. The third cluster consisted of 24 European genotypes, together with 5 from Asia and 1 from America, while 42 European cultivars were assigned to the fourth cluster, with 1 from Asia and 1 from America. The fourth cluster mainly contained genotypes bred in Western (British, French) and Central Europe (Germany, Austria, Switzerland). There was a higher percentage of Hungarian lines in the second and third clusters (59% and 32%), while they made up only 10.5% of the first and 14.6% of the fourth cluster. Cultivars from Eastern and South Eastern Europe were found with greater frequency in clusters 2 and 3 (18.5% and 38.7%), which could be related to the fact that genotypes from these regions serve as crossing material in each other’s breeding programmes. The degree of diversity within the four clusters was fairly similar and quite high; only in cluster 2 could distinct subgroups be detected (Fig. 1). A comparison of genetic distances revealed a medium negative significant correlation between the origin of the genotypes and the population structure (r = – 0 .54; P ≤ 0.001).
Allele distribution of major developmental genes
An analysis was made of the allele distribution frequency of the major plant development genes (VRN and PPD) in genotypes in each of the four clusters (Supplemental Table 1). The spring alleles of the VRN-A1, VRN-B1 and VRN-D1 genes were present in 30%, 54% and 17% of the genotypes, respectively, in the first cluster, while these figures were 2%, 7% and 9% in cluster 2, 3%, 14% and 7% in cluster 3 and 14%, 11% and 0% in cluster 4. The photoperiod-insensitive allele types of PPD-D1 and PPD-B1 were carried by 43% and 30%, respectively, of the genotypes in cluster 1, while these ratios were 55% and 11% for cluster 2, 86% and 58% for cluster 3 and 25% and 13% for cluster 4. The distribution of the copy number of the PPD-B1 gene in the genotypes was examined within each cluster, and the distribution of the intercopy structure of this gene detected in ‘Chinese Spring’ was also investigated. In the first and third clusters, 30% and 58%, respectively, of the genotypes carried more than one copy of this gene, while these figures were only 9% and 14%, respectively, for clusters 2 and 4. In the third cluster, the ‘Chinese Spring’ intercopy structure was observed in 59% of the genotypes, while it was present in 7% of the first cluster and 5% of the second. This allele could not be detected in wheat genotypes in cluster 4. The lower ratio of the spring alleles in the three VRN1 genes together with the higher ratio of photoperiod sensitivity alleles in the two PPD1 genes could explain the later heading date of genotypes in the fourth cluster.
The genotypic results were in agreement with phenotypic observations, as the mean heading dates determined for the genotypes in each cluster showed that, averaged over three years, plants in clusters 1, 2 and 3 reached this phenophase more rapidly than those in cluster 4. This is illustrated in terms of the mean values of the effective heat sum (ETT) required to reach the Z59 stage in Fig. 3.
Discussion
The results of genetic diversity studies showed that the tested cultivars could be divided into four clusters, primarily on the basis of geographical origin. The first cluster consisted mainly of American and Asian cultivars, cluster 2 of Central European, cluster 3 of Hungarian and South European and cluster 4 of West European cultivars. The cultivars used in breeding programmes in Central and South Eastern Europe were found in all four clusters, confirming the findings of other authors, who reported that the wheat genotypes in this region of Europe had greater genetic diversity than their Western and Northern European counterparts (Roussel et al. 2005; Hai et al. 2007; Zhang et al. 2011; Novoselović et al. 2016; El-Esawi et al. 2018). This could be attributed to the diverse environmental and soil conditions and to differences in breeding techniques (Roussel et al. 2005). The isolation caused by the Alps and the Carpathians also played an important role in the separation of relationships between Western and South Eastern genotypes (Roussel et al. 2005). The great genetic diversity observed in Hungarian breeding materials could partly be explained by the unusual climatic conditions in Hungary, which is located at the meeting point of three climatic zones, the Oceanic (cool summer, mild winter, small annual heat fluctuation, uniform precipitation distribution), the Continental (warm summer, cold, dry winter, large annual heat fluctuation, precipitation maxima in early summer and autumn) and the Mediterranean (hot, dry summer, mild winter, precipitation maxima in autumn and winter). Each year the weather components of these three zones are experienced with different intensity and frequency, and very often in mixed or overlapping forms, thus exerting a high level of selection pressure during breeding. This means that only lines with a combination of genes and alleles that makes them capable of adapting to such diverse conditions become state-registered cultivars.
In setting up this multi-varietal wheat population based on the heading date characteristics, our aim was to study the genetic diversities in wheats with winter growth habit. Thus it is interesting to note that relatively high numbers of spring alleles were identified in the three Vrn1 vernalisation genes. There were however remarkable differences in the ratio of spring alleles between the four clusters, which were primarily found in the American and Asian cultivars and were thus present in a much higher ratio in cluster 1 than in the other clusters. The photoperiod-sensitive alleles of the PPD-B1 and PPD-D1 genes were detected in a much greater number in samples assigned to cluster 4. A close correlation was revealed between this recessive allele type and later heading date (Laurie et al. 1995; Turner et al. 2005; Beales et al. 2007; Díaz et al. 2012; Kiss et al. 2014). The genotypes in clusters 1 and 3, on the other hand, contained a higher proportion of the photoperiod-insensitive alleles of the PPD1 genes, which led to a substantial reduction in the time required to reach the Z59 phenophase. These two clusters also contained a higher ratio of genotypes carrying more than one copy of the PPD-B1 gene. The absence of the ‘Chinese Spring’ intercopy structure of this gene from the genotypes in cluster 4 was also correlated with later heading, as the point mutation detected in the Chinese Spring cultivar exhibits cosegregation with the early heading phenotype (Beales et al. 2007; Díaz et al. 2012; Kiss et al. 2014).
One precondition for successful breeding is the presence of a satisfactory level of genetic diversity, which promotes the chances that favourable properties will be manifested, thus ensuring the selection of lines with better adaptability. For this reason, knowledge of the extent of genetic diversity, the allele distribution of the major plant development genes influencing yield potential and their relationship with the phenotype of breeding lines could provide breeders with extremely important information.
References
Almanza-Pinzón MI, Khairallah M, Fox PN, Warburton ML (2003) Comparison of molecular markers and coefficients of parentage for the analysis of genetic diversity among spring bread wheat accessions. Euphytica 130:77–86
Andeden EE, Yediay FE, Baloch FS, Shaaf S, Nachit KB, M, Özkan H, (2011) Distribution of vernalization and photoperiod genes (Vrn-A1, Vrn-B1, Vrn-D1, Vrn-B3, Ppd-D1) in turkish bread wheat cultivars and landraces. Cer Res Commun 39:352–364
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimising parental selection for genetic linkage maps. Genome 36:181–186
Astarini AI, Plummer AJ, Lancaster AR, Yan G (2004) Fingerprinting of cauliflower cultivars using RAPD markers. Aust J Agric Res 55:117–124
Balfourier F, Roussel V, Strelchenko P, Exbrayat FV, Sourdille P, Boutet G, Koenig J, Ravel C, Mitrofanova O, Beckert M, Charment G (2007) A worldwide bread wheat core collection arrayed in a 384-well plate. Theor Appl Genet 114:1265–1275
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733
Benson J, Brown-Guedira G, Murphy J, Sneller C (2012) Population structure, linkage disequilibrium, and genetic diversity in soft winter wheat enriched for fusarium head blight resistance. Plant Gen 5:71–80
Blake NK, Lanning SP, Martin JM, Doyle M, Sherman JD, Naruoka Y, Talbert LE (2009) Effect of variation for major growth habit genes on maturity and yield in five spring wheat populations. Crop Sci 49:1211–1220
Bogard M, Pierre JB, Huguenin-Bizot B, Hourcade D, Paux E, Le Bris X, Gouache D (2015) A simple approach to predict growth stages in winter wheat (Triticum aestivum L.) combining prediction of a crop model and marker based prediction of the deviation to a reference cultivar: a case study in France. Eur J Agron 68:57–68
Börner A, Worland AJ, Plaschke J, Schumann E, Law CN (1993) Pleiotropic effects of genes for reduced height (Rht) and day-length insensitivity (Ppd) on yield and its componentsfor wheat grown in middle Europe. Plant Breed 111:204–216
Borràs G, Romagosa I, van Eeuwijk F, Slafer GA (2009) Genetic variability in duration of pre-heading phases and relationships with leaf appearance and tillering dynamics in a barley population. Field Crops Res 113:95–104
Bouffier L, Raffin A, Kremer A (2008) Evolution of genetic variation for selected traits in successive breeding populations of maritime pine. Heredity 101:156–165
Chao S, Zhang W, Akhunov E, Sherman J, Ma Y, Luo MC, Dubcovsky J (2008) Analysis of gene-derived SNP marker polymorphism in US wheat (Triticum aestivum L.) cultivars. Mol Breed 23:23–33
Charmet G (2011) Wheat domestication: lessons for the future. C R Biol 334:212–220
Chen Y, Carver BF, Wang S, Zhang F, Yan L (2009) Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor Appl Genet 118:881–889
Chen Y, Carver BF, Wang S, Cao S, Yan L (2010) Genetic regulation of developmental phases in winter wheat. Mol Breed 26:573–582
Crossa J, Burgueno J, Dreisigacker S, Vargas M, Herrera-Foessel SA, Lillemo M, Singh RP, Trethovan R, Warburton M, Franco J, Reynolds M, Crouch JH, Ortiz R (2007) Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177:1889–1913
Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA (2012) Copy number variation affecting the photoperiod-B1 and vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE 7(3):e33234
Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G (1998) Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet 97:968–975
El-basyoni I, Baenziger PS, Dweikat I, Wang D, Eskridge K, Saadalla M (2013) Using DArT markers to monitor genetic diversity throughout selection: a case study in Nebraska’s winter wheat breeding nurseries. Crop Sci 53:2363–2373
El-Esawi AM, Witczak J, El-Fatah AA, Ali HM, Elshikh MS, Ahmed M (2018) Analysis of the genetic diversity and population structure of Austrian and Belgian wheat germplasm within a regional context based on DArT markers. Genes 9:47
Faure S, Higgins J, Turner A, Laurie DA (2007) The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare L.). Genetics 176:599–609
Ficco PDBM, Russo MA, De Vita P, Papa R, Rubiales D (2012) Characterization of wheat DArT markers: genetic and functional features. Mol Gen Genom 287:741–753
Fu D, Szűcs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Gen Genom 273:54–65
Fu YB, Peterson GW, Yu JK, Gao L, Jia J, Richards KW (2006) Impact of plant breeding on genetic diversity of the Canadian hard red spring wheat germplasm as revealed by ESTderived SSR markers. Theor Appl Genet 112:1239–1247
Fufa H, Baenziger PS, Beecher BS, Dweikat I, Graybosch RA, Eskridge KM (2005) Comparison of phenotypic and molecular marker-based classifications of hard red winter wheat cultivars. Euphytica 145:133–146
González FG, Slafer GA, Miralles DJ (2005) Photoperiod during stem elongation in wheat: Is its impact on fertile floret and grain number determination similar to that of radiation? Funct Plant Biol 32:181–188
Hai L, Wagner C, Friedt W (2007) Quantitative structure analysis of genetic diversity among spring bread wheats (Triticum aestivum L.) from different geographical regions. Genetica 130:213–225
Huang XQ, Börner A, Röder MS, Ganal MW (2002) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor Appl Genet 105:699–707
Huang XQ, Wolf M, Ganal MW, Orford S, Koebner RMD, Röder MS (2007) Did modern plant breeding lead to genetic erosion in European winter wheat varieties? Crop Sci 47:343–349
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucl Acids Res 29:E25
Kabbaj H, Sall AT, Al-Abdallat A, Geleta M, Amri A, Filali-Maltouf A, Belkadi B, Ortiz R, Bassi FM (2017) Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Front Plant Sci 8:1277
Karsai I, Gy V, Petrovics S, Petcu E, Kobiljski B, Ivanovska S, Bedő Z, Veisz O (2012) Assessment of the spatial genotypic and phenotypic diversity present in the various winter wheat breeding programs in Southeast Europe. Euphytica 186:139–151
Kato H, Taketa S, Ban T, Iriki N, Murai K (2001) The influence of a spring habit gene, Vrn-D1, on heading time in wheat. Plant Breeding 120:115–120
Khan IA, Awan FS, Ahmad A, Fu YB, Iqbal A (2005) Genetic diversity of Pakistan wheat germplasm as revealed by RAPD markers. Genet Resour Crop Evol 52:239–244
Kiss T, Balla K, Veisz O, Láng L, Bedő Z, Griffiths S, Isaac P, Karsai I (2014) Allele frequencies in the VRN-A1, VRN-B1, VRN-D1 vernalization response and PPD-B1 and PPD-D1 photoperiod sensitivity genes, and their effects on heading in a diverse set of wheat cultivars (Triticum aestivum L.). Mol Breed 34:297–310
Laurie DA, Pratchett N, Bezant JH, Snape JW (1995) RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter 9 spring barley (Hordeum vulgare L) cross. Genome 38:575–585
Law CN, Sutka J, Worland AJ (1978) A genetic study of day-length response in wheat. Heredity 41:575–585
Lopes MS, El-Basyoni I, Baenziger PS, Singh S, Royo C, Ozbek K, Aktas H, Ozer E, Ozdemir F, Manickavelu A, Ban T, Vikram P (2015) Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J Exp Bot 66(12):3477–3486
Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J (2005) Regulation of VRN-1 vernalization response genes in normal and transgenic polyploid wheat. Plant Physiol 138:2364–2373
Matthies IE, Hintum T, Weise S, Röder M (2012) Population structure revealed by different marker types (SSR or DArT) has an impact on the results of genome-wide association mapping in European barley cultivars. Mol Breed 30:951–966
Mukhtar MS, Rahman M, Zafar Y (2002) Assessment of genetic diversity among wheat (Triticum aestivum L.) cultivars from a range of localities across Pakistan using random amplified polymorphic DNA (RAPD) analysis. Euphytica 128:417–425
Nielsen NH, Backes G, Stougaard J, Andersen SU, Jahoor A (2014) Genetic diversity and population structure analysis of European hexaploid bread wheat (Triticum aestivum L.) varieties. PLoS ONE 9(4):94000
Novoselović D, Bentley AR, Šimek R, Dvojković K, Sorrells ME, Gosman N, Horsnell R, Drezner G, Šatović Z (2016) Characterizing Croatian wheat germplasm diversity and structure in a European context by DArT markers. Front Plant Sci 7:184
Orabi J, Jahoor A, Backes G (2014) Changes in allelic frequency over time in European bread wheat (Triticum aestivum L.) varieties revealed using DArT and SSR markers. Euphytica 197:447–462
Parker GD, Fox PN, Langridge P, Chalmers K, Whan B, Ganter PF (2002) Genetic diversity within Australian wheat breeding programs based on molecular and pedigree data. Euphytica 124:293–306
Pritchard JK, Stephens M, Donnelly PJ (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pugsley AT (1971) A genetic analysis of the spring-winter habit of growth in wheat. Aust J Agric Res 22:21–23
Pugsley AT (1972) Additional genes inhibiting winter habit in wheat. Euphytica 21:547–552
Raman H, Stodart BJ, Cavanagh C, Mackay M, Morell M, Milgate A, Martin P (2010) Molecular diversity and genetic structure of modern and traditional landrace cultivars of wheat (Triticum aestivum L.). Crop Pasture Sci 61:222–229
Rauf S, Khan AA, Naveed A, de Silva T (2010) Consequences of plant breeding on genetic diversity. Int J Plant Breed 4:1–21
Reif JC, Zhang P, Dreisigacker S, Warburton ML, van Ginkel M, Hoisington D, Bohn M, Melchinger AE (2005) Wheat genetic diversity trends during domestication and breeding. Theor Appl Genet 110:859–864
Röder MS, Wendehake K, Korzun V, Bredemeijer G, Laborie D, Bertrand L, Isaac P, Rendell S, Jackson J, Cooke RJ, Vosman B, Ganal MW (2002) Construction and analysis of a microsatellite-based database of European wheat varieties. Theor Appl Genet 106:67–73
Roussel V, Leisova L, Exbrayat F, Stehno Z, Balfourier F (2005) SSR allelic diversity changes in 480 european bread wheat varieties released from 1840 to 2000. Theor Appl Genet 111:162–170
Semagn K, Bjornstad A, Skinnes H, Maroy AG, Tarkegne Y, William M (2006) Distribution of DArT, AFLP, and SSR markers in a genetic linkage map of a doubled-haploid hexaploid wheat population. Genome 49:545–555
Siedler H, Messmer MM, Schachermayr GM, Winzeler H, Winzeler M, Keller B (1994) Genetic diversity in European wheat and spelt breeding material based on RFLP data. Theor Appl Genet 88:994–1003
Smale M (1997) The green revolution and wheat genetic diversity: some unfounded assumptions. World Dev 25:1257–1269
Snape JW, Law CN, Parker BB, Worland AJ (1985) Genetical analysis of chromosome 5A of wheat and its influence on important agronomic characters. Theor Appl Genet 71:518–526
Spanic V, Korzun V, Ebmeyer E (2016) Assessing genetic diversity of wheat genotypes from different origins by SNP markers. Cer Res Commun 44(3):361–369
Stachel M, Lelley T, Grausgruber H, Vollmann J (2000) Application of microsatellites in wheat (Triticum aestivum L.) for studying genetic differentiation caused by selection for adaptation and use. Theor Appl Genet 100:242–248
Stodart BJ, Mackay M, Raman H (2005) AFLP and SSR analysis of genetic diversity among landraces of bread wheat (Triticum aestivum L. em. Thell) from different geographic regions. Aust J Agric Res 56:691–697
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327:818–822
Tottman DR, Makepeace RJ (1979) An explanation of the decimal code for the growth stages of cereals, with illustrations. Ann Appl Biol 93:221–234
Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310:1031–1034
van de Wouw M, van Hintum T, Kik C, van Treuren R, Visser B (2010) Genetic diversity trends in twentieth century crop cultivars: a meta analysis. Theor Appl Genet 120:1241–1252
White J, Law JR, MacKay I, Chalmers KJ, Smith JSC, Kilian A, Powell W (2008) The genetic diversity of UK, US and Australian cultivars of Triticum aestivum measured by DArT markers and considered by genome. Theor Appl Genet 116:439–453
Worland AJ (1996) The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 89:49–57
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109:1677–1686
Yang FP, Zhang XK, Xia XC, Laurie DA, Yang WX, He ZH (2009) Distribution of the photoperiod insensitive Ppd-D1a allele in Chinese wheat cultivars. Euphytica 165:445–452
Zhang LY, Liu D, Guo X, Yang WL, Sun JZ, Wang DW, Sourdille P, Zhang AM (2011) Investigation of genetic diversity and population structure of common wheat cultivars in northern China using DArT markers. BMC Genet 12:42
Acknowledgments
This research was funded by research grants from the Hungarian Scientific Research Fund (NKFIH-FK-134234, NKFIH-K-129221 and NKFIH-K-119801).
Funding
Open Access funding provided by Eszterhazy Karoly University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.K. Khlestkina.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kiss, T., Balla, K., Cseh, A. et al. Assessment of the genetic diversity, population structure and allele distribution of major plant development genes in bread wheat cultivars using DArT and gene-specific markers. CEREAL RESEARCH COMMUNICATIONS 49, 549–557 (2021). https://doi.org/10.1007/s42976-021-00136-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-021-00136-2