Introduction
The olive fruit fly Bactrocera oleae (Rossi) (Diptera, Tephritidae) causes enormous losses in olive production and is present in the great majority of olive orchards in the world. The total loss in olive production has been estimated to up to 30% of the production (Bueno and Jones, Reference Bueno and Jones2002; Kampouraki et al., Reference Kampouraki, Stavrakaki, Karataraki, Katsikogiannis, Pitika, Varikou, Vlachaki, Chrysargyris, Malandraki, Sidiropoulos, Paraskevopoulos, Gilpathi, Roditakis and Vontas2018). Current control measures against olive fly rely on the use of chemical insecticides; primarily bait applications targeting the adult fly. However, insecticide resistance substantially affects the efficiency of control applications in some regions (Vontas et al., Reference Vontas, Hernández-Crespo, Margaritopoulos, Ortego, Feng, Mathiopoulos and Hsu2011; Kampouraki et al., Reference Kampouraki, Stavrakaki, Karataraki, Katsikogiannis, Pitika, Varikou, Vlachaki, Chrysargyris, Malandraki, Sidiropoulos, Paraskevopoulos, Gilpathi, Roditakis and Vontas2018). Alternative control methods have been developed and/or tried such as biological control, the Sterile Insect Technique (SIT), mass trapping and the use of genetics-enhanced SIT. However, they have not produced very encouraging results due to either their higher cost and/or their low effectiveness against high infestations (Raspi et al., Reference Raspi, Loni, Canovai and Canale2007; Daane and Johnson, Reference Daane and Johnson2010; Ant et al., Reference Ant, Koukidou, Rempoulakis, Gong, Economopoulos, Vontas and Alphey2012; Nestel et al., Reference Nestel, Rempoulakis, Yanovski, Miranda, Papadopoulos, Rami Horowitz and Ishaaya2016).
A possible alternative target is the symbiotic bacterium Candidatus Erwinia dacicola which is required for the development of the larvae in the olive, especially in the un-ripe green olives (Ben-Yosef et al., Reference Ben-Yosef, Pasternak, Jurkevitch and Yuval2015). Recently, the interruption of symbiosis in the olive fruit fly was targeted in pilot trials by copper and other formulations with some promising results (Bigiotti et al., Reference Bigiotti, Pastorelli, Guidi, Belcari and Sacchetti2019; Sinno et al., Reference Sinno, Bézier, Vinale, Giron, Laudonia, Garonna and Pennacchio2020). In fact, targeting the symbiont (‘dysbiosis’) may open the way for novel classical or biotechnology-based environmentally friendly strategies in olive pest control.
The symbiont has been identified in various organs of the fly, it is present in all life stages (Estes et al., Reference Estes, Hearn, Bronstein and Pierson2009) and it is suggested to be transmitted from the female fly to the egg via the ovipositor (Estes et al., Reference Estes, Hearn, Bronstein and Pierson2009). The role of the bacterium is not entirely understood but research has shown that the bacteria enhanced egg-production of the female when fed on a nitrogen-poor diet (Ben-Yosef et al., Reference Ben-Yosef, Pasternak, Jurkevitch and Yuval2014). Importantly, they are strictly required for larval development in green un-ripe olives (Ben-Yosef et al., Reference Ben-Yosef, Aharon, Jurkevitch and Yuval2010), possibly for detoxifying defensive plant phenolic compounds (Soler-Rivas et al., Reference Soler-Rivas, Epspin and Wichers2000) and/or sequestering amino acids for protein synthesis (Ben-Yosef et al., Reference Ben-Yosef, Pasternak, Jurkevitch and Yuval2014, Reference Ben-Yosef, Pasternak, Jurkevitch and Yuval2015).
The genome of the bacterium has been sequenced (Blow et al., Reference Blow, Gioti, Starns, Ben-Yosef, Pasternak, Jurkevitch, Vontas and Darby2016; Estes et al., Reference Estes, Hearn, Agrawal, Pierson and Dunning Hotopp2018a, Reference Estes, Hearn, Nadendla, Pierson and Dunning Hotoppb; Blow et al., Reference Blow, Gioti, Goodhead, Kalyva, Kampouraki, Vontas and Darby2020) and a transcriptomics analysis comparing bacteria form larvae reared on either green or black olives has been carried out (Pavlidi et al., Reference Pavlidi, Gioti, Wybouw, Dermauw, Ben-Yosef, Yuval, Jurkevich, Kampouraki, Van Leeuwen and Vontas2017).
Ca. E. dacicola cannot be cultured in vitro and thus the study of these bacteria is difficult, as they have to be obtained from wild-caught flies which are only present during restricted periods of the year. Furthermore, established laboratory strains raised on the artificial diet are devoid of the symbiont due to the use of antibiotics during many years of rearing (Estes et al., Reference Estes, Hearn, Burrack, Rempoulakis and Pierson2012a). Previous experience has revealed that establishing populations from wild flies is a difficult and time-consuming endeavor (Estes et al., Reference Estes, Nestel, Belcari, Jessup, Rempoulakis and Economopoulos2012b; Koskinioti et al., Reference Koskinioti, Ras, Augustinos, Tsiamis, Beukeboom, Caceres and Bourtzis2019). Methods to transfer the bacterium from wild-caught flies to laboratory flies would thus greatly assist in its study. Attempts in this direction have suggested that cohabitation of infected and non-infected flies results in cross-infection (Estes et al., Reference Estes, Segura, Jessup, Wornoayporn and Pierson2014; Bigiotti et al., Reference Bigiotti, Pastorelli, Guidi, Belcari and Sacchetti2019).
Here we describe a robust method for transfer of the bacteria from wild flies to a laboratory strain. This method will allow controlled studies of the interaction between the symbiotic bacteria and the insect host, towards the development of innovative ‘dysbiosis’-based pest control strategies.
Materials and methods
B. oleae strains
The laboratory strain of B. oleae was obtained from National Centre of Scientific Research Democritos (Athens, Greece) in 2001. The strain was routinely treated with antibiotics and is devoid of Ca. E. dacicola (Estes et al., Reference Estes, Hearn, Burrack, Rempoulakis and Pierson2012a).
Third instar larvae were sampled from infested green olives collected from olive orchards in Prefecture of Heraklion, Crete, Greece; 35°18′18.47″N 25°04′25.9″E, 82 m above sea level, FORTH campus and 35°19′19.8″N 25°09′26.0″E, 79 m above sea level, Agios Ioannis, both located within Heraklion municipality, Greece in September 2015. The olives were kept at 24°C ± 1°C and third instar larvae were collected when they had exited the olives. Adults were collected for 3–4 h in McPhail traps with 2% ammonia as an attractant (food lure traps) from the same locations (September–November 2015) and were immediately used for the experiments.
Rearing of B. oleae
B. oleae strain Democritos was reared at 24°C ± 1°C at 50–60% humidity and with 16:8 h light:dark photoperiod in plexiglass cages (size 100 × 30 × 30 cm3). Females were allowed to oviposit on ceresine cones. Eggs were not treated with antifungal agents. Newly hatched larvae were transferred onto the surface of larval medium. At the end of the larval stage the emerging larvae were transferred to Petri dishes containing sawdust for pupation. Larval and adult food was as described by Tzanakakis (Reference Tzanakakis and Hooper1989); during the last 15 years antibiotics is not added to the food. Samples intended for polymerase chain reaction (PCR) were snap-frozen in liquid nitrogen and stored at −80°C.
Cross of laboratory strain to wild flies
Wild male or female flies were collected in McPhail bait traps positioned in infested orchards and mixed with Democritus flies of the opposite gender. In cages with dimensions 11 × 9.5 × 8 cm3 one virgin female was positioned together with three males of the opposite strain. Wild females were provided with fresh olives for egg-laying. Democritos females were allowed to oviposit in ceresine cones. Progeny was tested for the presence of bacteria. DNA was extracted from pools of four third instar F1 larvae from both crosses and diagnostic PCR was performed.
Cohabitation of wild and laboratory populations
In total, 200 male and female Democritos flies were mixed with 35 wild males in a big cage (size 100 × 30 × 30 cm3). The females were allowed to lay eggs and the resulting F1 males flies were mixed with females of the Democritos strain and wild males. The resulting F2 flies were inbred and the F3 generation was tested for the presence of the bacteria by diagnostic PCR.
Incubation of eggs with gastric caeca from wild flies
Six gastric caeca were dissected from larvae of wild flies and gently squashed before incubation on wet filter paper with about 100 newly laid eggs of the Democritos population for 30 min. The eggs were then transferred to larval food and allowed to develop to third instar larvae. Half were used for diagnostic PCR. Of the other half, gastric caeca were dissected and examined under the microscope for the presence of bacteria. The experiment was repeated once more.
Feeding laboratory flies with esophageal bulbs from wild flies
About 200 Democritos flies in a big plexiglass cage (size 100 × 30 × 30 cm3) were starved of food and water for 48 h. In total, 20 esophageal bulbs were dissected from adult wild flies. Ten of these were mixed with a drop of phosphate buffered saline (PBS) and positioned on a petri dish. The other ten were placed on wet filter paper. The dishes were added to the cage and offered as the only source of food and water for 24 h, after which time standard food and water was added to the cage. The females were allowed to lay eggs and F1 flies were tested by diagnostic PCR; F2 flies were also tested.
Transplantations of Ca. E. dacicola
Larval gastric caeca or adult esophageal bulbs were dissected using forceps from wild flies under the stereoscope in aseptic conditions. Whole organs were transplanted to adult female Democritos flies, anaesthetized by carbon dioxide, using an entomological needle (Black Anodized Insect Pins No 26000 size 000, from Fine Science Tools). The transplantation targeted head, thorax and abdomen. To establish single female families each female was mated to four Democritos males using the same conditions as described for the genetic crosses. The presence of bacteria was tested by diagnostic PCR.
DNA preparation
Genomic DNA was extracted using the CTAB protocol (Doyle, Reference Doyle1990). Insects were ground with plastic pestles in 50 μl of extraction buffer per fly (2% CTAB (Sigma), 100 mM Tris-HCl pH 8.0, 20 mM EDTA pH 8.0, 1.4 M NaCl, 0.2% 2-mercaptoethanol). The suspension was incubated for 15 minutes at 65°C, treated with RNase A for 1 h at 37°C, extracted with chloroform-isoamyl alcohol and DNA was precipitated with isopropanol.
Diagnostic PCR
Diagnostic PCR to detect the presence of Erwinia was performed on genomic DNA prepared from pools of flies or individuals. All samples were tested with primers targeting the fly genomic β-actin gene primers (5′-CGGTATCCACGAAACCACAT-3′ and 5′-ATTGTTGATGGAGCCAAAGC-3′) to confirm the quality of the DNA preparation. To test for Ca. E. dacicola PCR was carried out using the following primer pair specific for the recA gene.: 5′-TCTACCGGTTCGTTATCCCTG-3′ and 5′- AACGATAATCACGTCAACAGCAC-3′ and for the S16 gene 5′-GAAGGCGAAGAGGTTAATAACCTTTTT-3′ and 5′-AGGCGCTCTCCCGAAGTTAAG-3′ The primers were designed using the transcriptome of Ca. E. dacicola (Pavlidi et al., Reference Pavlidi, Gioti, Wybouw, Dermauw, Ben-Yosef, Yuval, Jurkevich, Kampouraki, Van Leeuwen and Vontas2017). They were tested extensively with samples of laboratory flies and wild flies to determine their specificity for Ca. E. dacicola. KapaTaq Polymerase (Roche) was used for all reactions and cycling conditions were: initial denaturation of 95°C for 3 min followed by 35 cycles of 95°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 45 s, and a final extension at 72°C for 10 min. Reactions were run on 1.5% agarose gels and imaged in a gel documentation system (BioRad).
Results and discussion
In order to carry out detailed studies of the interaction of Ca. E. dacicola with B. oleae controlled infections are required. The Democritos laboratory strain, established many years ago, was initially reared in the presence of antibiotics and is therefore devoid of Ca. E. dacicola (Estes et al., Reference Estes, Hearn, Burrack, Rempoulakis and Pierson2012a). We aimed to develop a robust method to transfer bacteria from wild flies to the Democritos laboratory strain.
Unsuccessful approaches for transfer of Ca. E. dacicola from wild flies to the laboratory population
Cross between laboratory strain and wild flies
In our first attempt, we crossed wild flies with the laboratory population reasoning that possibly bacteria could be transferred from wild males to Democritos females. The reciprocal crosses were carried out; Democritos females were crossed to wild males and wild females were crossed to laboratory males. In the latter case, the females were allowed to lay eggs in olives. PCR using primers specific for Ca. E. dacicola were used to determine the presence of the bacteria. The results (fig. 1A, B) showed that Ca. E. dacicola is only transferred from females to the progeny. Thus it is difficult to transfer the bacteria from wild flies as in our experience field females do not oviposit in an artificial medium.
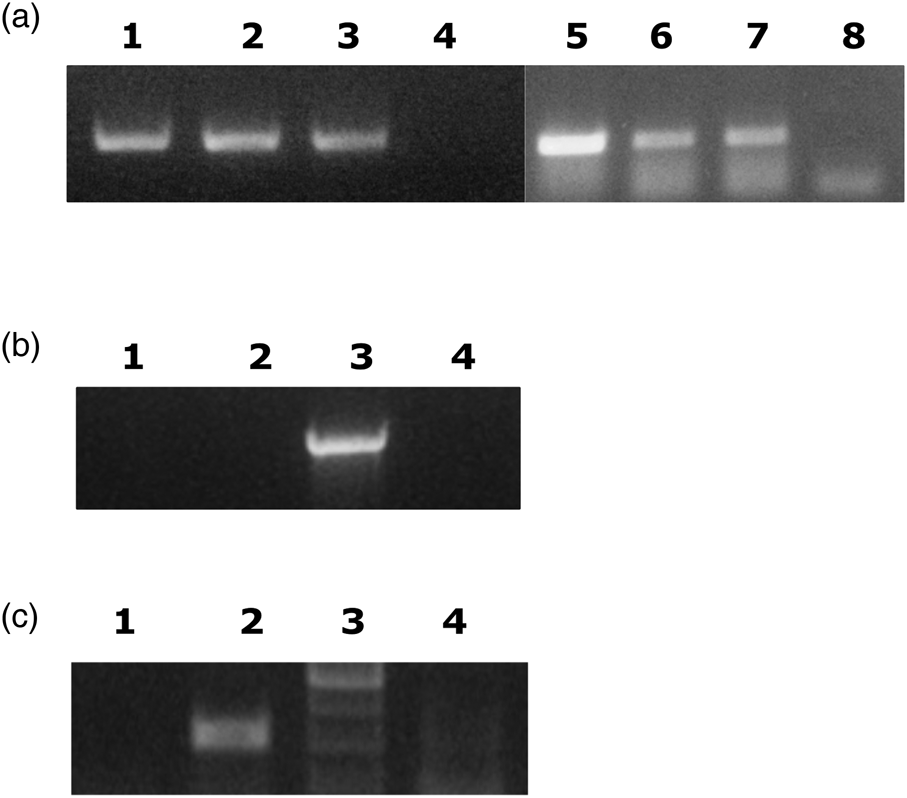
Figure 1. Ca. E. dacicola is only transferred to the next generation by females. A. Diagnostic PCR of individual F1 progeny from two crosses of WT females with Democritos males (Lane 1, 2, 5–7). Lane 3; positive control (larvae of wild flies). Lanes 4 and 8; negative controls, no template added. Primers targeted recA of Ca. E. dacicola. B. Diagnostic PCR shows the absence of bacteria in the progeny from a cross of wild males with Democritos females. DNA was prepared from pools of four third instar larvae. Two independent samples are shown (lanes 1 and 2). Positive control as in A (lane 3). Lane 4; no DNA template added. Primers targeted the Ca. E. dacicola 16S gene. C. Diagnostic PCR of the F3 generation from cohabitation of wild-caught males with males and females of the Democritos strain. DNA was prepared from pools of four third instar larvae. Results from one pool are shown. Lane 1; F3 generation of flies from cohabitation. Lane 2; positive control as in A. Lane 3, molecular weight marker. Lane 4; negative control (no DNA added).
Cohabitation of wild and laboratory populations
It has previously been reported that cohabitation of a laboratory population lacking the symbiont with males of a wild population resulted in the transfer of the symbiont to the laboratory flies (Estes et al., Reference Estes, Segura, Jessup, Wornoayporn and Pierson2014; Bigiotti et al., Reference Bigiotti, Pastorelli, Guidi, Belcari and Sacchetti2019). We repeated these experiments by mixing males from the field with Democritos males and females and testing whether the bacteria were transferred to the next generation. The resulting F1 males from the mixed population were kept in the same cage as laboratory males and females. The F2 population was inbred and samples from the F3 generation were tested by diagnostic PCR. All samples were negative for the presence of Ca. E. dacicola (one example is showed in fig. 1C). Our experiments suggest that cohabitation of wild males with laboratory females does not lead to stable infection of the laboratory flies. This is in contrast to previous studies that suggested that horizontal transfer of the bacteria indeed takes place during cohabitation (Estes et al., Reference Estes, Segura, Jessup, Wornoayporn and Pierson2014; Bigiotti et al., Reference Bigiotti, Pastorelli, Guidi, Belcari and Sacchetti2019). Estes et al. recovered PCR fragments specific for Ca. E. dacicola from a laboratory population derived from mating of the Democritos strain with wild males; the offspring had been maintained for ~20 generations. In the paper by Bigiotti et al. only the flies from the experiment were tested but not the offspring, summing up these conflicting results horizontal transfer may take place in certain circumstances but may not be easily reproduced in other laboratories.
Incubation of eggs with gastric caeca from wild flies
We next tested if we could transfer the bacteria to Democritos flies by providing Ca. E. dacicola obtained by dissection of gastric caeca from wild larvae. These were obtained from infested olives. Several methods were tested. In the first series of experiments, gastric caeca were dissected from the larvae, gently ruptured and then added to eggs oviposited by Democritos females. The experiment was repeated once with different numbers of eggs and gastric caeca. The larvae that developed from the eggs were tested by diagnostic PCR for the presence of the bacteria but no bacteria were detected (fig. 2A, table 1).

Figure 2. Attempted transfer of Ca. E. dacicola from wild flies to Democritos strain. A. Incubation of eggs with gastric caeca followed by diagnostic PCR of DNA extracted from the resulting third instar larvae. 100 newly laid eggs were incubated with dissected gastric caeca on wet filter paper for 30 min before moving the eggs to larval food. Pools of four larvae were extracted and two pools are shown (lanes1 and 2). DNA isolated from larvae of the wild population was used as a positive control (lane 3). Lane 4; no DNA template added. The recA primers were used for the PCR.
Table 1. Attempts to transfer Ca. E. dacicola from wild flies to Democritos laboratory strain.

nt, not tested. Pools of flies were tested.
Feeding laboratory flies with esophageal bulbs from wild flies
In the next experiment, we tested the transfer of the bacteria by feeding larvae of the laboratory population with esophageal bulbs from adult wild flies. The bulbs were either provided in a drop of PBS or on a filter paper soaked with water. The larvae were allowed to develop and resulting adults were mated to each other. Individuals from the F1 and F2 generations were tested by PCR but no infection was detected in any sample (table 1).
The results of these experiments suggest that stable transfer of the bacteria does not take place if they are provided to the insect in the food or to the eggs. One possibility is that the fly only takes up the bacterium during specific stages during its life cycle for example when the larva emerges from the embryo. Another explanation is that the bacteria are not viable after exposure to air. As has been suggested previously (Bigiotti et al., Reference Bigiotti, Pastorelli, Guidi, Belcari and Sacchetti2019) it is quite possible that Ca. E. dacicola that live inside the intestine of the fly is microaerophilic and does not tolerate high levels of oxygen. It is well known that this bacterium cannot be cultured though the reason for this to our knowledge has not been thoroughly investigated. Therefore, the exact reason for the lack of transfer in our case is still an open question.
Successful transfer of Ca. E. dacicola to the laboratory population by transplantation
Next, we tested transferring Ca. E. dacicola by transplantation from wild flies to the Democritos strain. In one experiment gastric caeca were dissected from larvae and transferred by transplantation to the abdomen of recipient adult females of the laboratory strain. In another series of experiments, esophageal bulbs of adult wild females were transplanted to the abdomen, thorax or head, respectively, of the recipient females. The surviving females were each mated with four males of the laboratory strain and eggs were collected. All surviving recipient females were tested by diagnostic PCR to determine if they carried Ca. E. dacicola (figs 3A and 4A, table 2). Positive recipient individuals were identified from three out of the four experiments. No positive fly was recovered from the flies transplanted with bacteria from the esophageal bulbs into the abdomen. Of the females receiving the transplant in the thorax, only two out of ten flies were positive but they were both sterile. On the other hand, several single female lines could be established from individuals transplanted with gastric caeca to the abdomen (two lines) and bulbs to the head (three lines). Adult individuals from these lines were tested by diagnostic PCR. The two lines from females transplanted with gastric caeca were followed until F3 and one individual tested was positive for Ca. E. dacicola (fig. 3B). Of the progeny of flies receiving bulbs seven F1 individuals originating from three mothers were also tested and they were all positive (fig. 4B). To strengthen these results, we repeated the transplantations of bulbs from adult wild flies to the head of females. Similar results were obtained as described above (table 2, Supplemental fig. S1).

Figure 3. Diagnostic PCR of B. oleae after transfer of bacteria from larval gastric caeca by transplantation. A. Progeny of three Democritos females after transplantation of larval gastric caeca to the abdomen. PCR was carried out on genomic DNA prepared from individual flies. Lanes 1–3 three daughters of recipient female #4, Lane 4, one daughter from female #6 and lane 5 and 6 two daughters to female #16. Lane 8 is an F2 progeny of recipient fly #16. The positive control used DNA derived from a wild fly (lane 7) while in the negative control (lane 9) DNA from Democritos flies was used as a template. B. F2 and F3 progeny of flies in A (lane 1–4). Lane 1 and 2 are F2 progeny of recipient fly #4, lane 3 from female #16 and lane 4 is an F3 individual from fly #16. Positive and negative control as in A (lane 5 and 6). Primers targeted Ca. E. dacicola recA.

Figure 4. Diagnostic PCR of B. oleae after transfer of bacteria from adult bulbs by transplantation. A. Democritos female recipients of transplantation of bulbs from adult wild flies to the head or abdomen. PCR was carried out with genomic DNA prepared from females transplanted to the head (lane 1–7, females H1, H2, H4–H8) and one transplanted to the abdomen (lane 8). The positive control used DNA derived from a wild-caught fly (lane 9) while in the negative control (lane 10) DNA from Democritos flies was used as a template. B. F1 progeny of single-pair matings derived from positive female's recipients after transplantation to the head. Two males and two females from the progeny of recipient female H5 (lanes 1–4), one male from female H6 (lane 5) and from female H8 (lane 6). Positive and negative control as in A (lane 7 and 8). Primers targeted recA of Ca. E. dacicola.
Table 2. Transplantation of Ca. E. dacicola from dissected organs of wild flies to Democritos laboratory strain.

Transplantation recipients were females in all cases.
Seven single female lines were established and the presence of bacteria was verified by diagnostic PCR in F3 individuals.
In conclusion, we show that it is possible to transmit Ca. E. dacicola by transplantation of bacteria derived from field samples into recipient laboratory female flies (summarized in fig. 5). The bacteria were vertically transmitted for at least three generations. Different target organs of the recipient flies were tested and our results suggest that transplantation of adult bulbs to the head may be the most efficient as a higher proportion of surviving recipients had received the bacterium and transmitted it to the F1 generation. However, numbers are small and more experiments are required to clarify this issue.

Figure 5. Graphic summary of transfer of symbiont by transplantation. Bacteria from larvae or adults derived from wild-caught samples (upper panel) were transferred by transplantation to adult laboratory female flies (lower panel). Different organs were targeted in the recipients as indicated. Transfers that resulted in stable transmission to the next generation are indicated by a solid arrow, whereas unsuccessful with a broken arrow.
This study, together with recent developments both in genomic resources (Estes et al., Reference Estes, Hearn, Nadendla, Pierson and Dunning Hotopp2018b; Bayega et al., Reference Bayega, Djambazian, Tsoumani, Gregoriou, Sagri, Drosopoulou, Mavragani-Tsipidou, Giorda, Tsiamis, Bourtzis, Oikonomopoulos, Dewar, Church, Papanicolaou, Mathiopoulos and Ragoussis2020) and genome modification technology (Koidou et al., Reference Koidou, Denecke, Ioannidis, Vlatakis, Livadaras and Vontas2020) may facilitate research to better understand the molecular basis of the olive fruit fly – Erwinia symbiosis (and possibly in other insect systems) in a systemic manner and utilize acquired knowledge towards the development of innovative control strategies that go beyond traditional approaches. The development of possible alternative efficient ‘dysbiosis’ approaches to control olive fly (Sinno et al., Reference Sinno, Bézier, Vinale, Giron, Laudonia, Garonna and Pennacchio2020) may prove a valid alternative, given the current and forthcoming strict EU regulations to reduce the use of neurotoxic insecticides. These could be based on effective and highly selective agents, safe for non-target organisms and non-vulnerable to insecticide resistance.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485321000031.
Acknowledgements
This research has been financed by Greek national funds through the Public Investments Program (PIP) of General Secretariat for Research & Technology (GSRT), under the Emblematic Αction ‘The Οlive Road’ (project code: 2018ΣΕ01300000). We also acknowledge the support of this work by the project ‘Advanced Research Activities in Biomedical and Agro alimentary Technologies’ (MIS 5002469) which is implemented under the ‘Action for the Strategic Development on the Research and Technological Sector,’ funded by the Operational Programme ‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014–2020, project code 2017ΣΕ1451000) and co-financed by Greece and the European Union (European Regional Development Fund).












