Abstract
A coronavirus (CoV) commonly known as SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) and causing COVID-19 (coronavirus disease of 2019) has become a pandemic following an outbreak in Wuhan. Although mutations in the SARS-CoV-2 spike glycoprotein (SGP) are obvious from comparative genome studies, the novel infectious nature of the virus, its new varients detected in the UK, and outside and recovery–death ratios of COVID-19 inspired us to review the mechanisms of the infection, replication, release, and transmission of progeny virions and the immune response in the host cell. In addition to the specificity of SARS-CoV-2 binding to angiotensin-converting enzyme 2 receptor and transmembrane protease serine 2, the varied symptoms and severity of the infection by the original and mutated forms of the virus suggest the significance of correlating the host innate and adaptive immunity with the binding of the virus to the mannose receptor via lipopolysaccharides (LPSs), toll-like receptors via LPS/proteins/RNA, and sialic acid (Sia) via hemagglutinin, or sugar-acid segments of glycans. HA-to-Sia binding is considered based on the innate Sia N-acetylneuraminic acid and the acquired Sia N-glycolylneuraminic acid in the epithelial cells and the sialidase/neuraminidase- or esterase-hydrolyzed release and transmission of CoVs. Furthermore, the cytokine storms common to aged humans infected with SARS-CoV-2 and aged macaques infected with SARS-CoV encourage us to articulate the mechanism by which the nuclear capsid protein and RNAs bypass the pattern recognition-induced secretion of interferons (IFNs), which stimulate IFN genes through the Janus-activated kinase-signal transducer and activator of a transcription pathway, leading to the secretion of antiviral proteins such as myxovirus resistance protein A/B. By considering the complexities of the structure, and the infectious nature of the virus and the structures and functions of the molecules involved in CoV infection, replication, and immune response, a new interface among virology, immunology, chemistry, imaging technology, drug delivery, and nanoscience is proposed and will be developed. This interface can be an essential platform for researchers, technologists, and physicians to collaborate and develop vaccines and medicines against COVID-19 and other pandemics in the future.
Similar content being viewed by others
Introduction
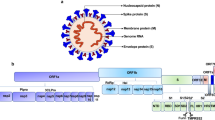
The halo or crown structure of the proteinaceous spike peplomers or glycoproteins (SGPs) detected in a transmission electron microscope image (Fig. 1) is the basis for the name coronavirus (CoV) given to viruses causing a series of respiratory illnesses, including COVID-19 (coronavirus disease of 2019), SARS (severe acute respiratory syndrome), and MERS (Middle East respiratory syndrome). The structure1, size (80–120 nm)2, genome3,4, and RNA-based pathogenesis5,6 of SARS-CoV-2 resemble those of other CoVs7. The highly pathogenic nature of SARS-CoV-2 and its recent genetic varients suggests that the binding affinities of these pathogens are high for a host cell, and it competently bypasses or blocks the cytokine (interferon (IFN))-triggered immune responses of a host cell. Thus, the following fundamental questions related to the tropism, replication, and release/transmission of SARS-CoV-2 face us. How does SARS-CoV-2 acclimate to the specific SGP genes3,4,7,8 that supplement the virus with a furin cleavage segment (FCS)1,9 to efficiently recognize angiotensin-converting enzyme 2 receptor (ACE2R)1,10 and bind to it? Is hemagglutinin (HA) a coreceptor for sialic acid (Sia)-based binding to a host cell? How does neuraminidase (NASe)- or esterase (ES)-based cleavage release a progeny virion? How do nonstructural proteins, the nuclear capsid (NC) and other structural proteins, and RNA bypass the IFN-induced JAK-STAT (Janus-activated kinase-signal transducer and activator of transcription) mechanism and form progeny virions11,12,13? Initially, SARS-CoV-2 was connected to Rhinolophus affinis (R. affinis), a bat species, because of the 96% sequence similarity between the 29.9 kb RNA of SARS-CoV-2 and the RNA of RaTG13 virus in R. affinis7,8,14. The similarities between the SGP amino acids of SARS-CoV-2 and Malayan pangolin (Manis javanica, M. javanica) CoV (pangolin-CoV) attracted further attention to the studies on the chimeric nature of the viral RNA and the origin of SARS-CoV-215,16. Independent of the suggested lineage relationships between RaTG13, SARS-CoV-2, and Pangolin-CoV, investigations have validated the zoonotic evolution of SARS-CoV-2. An emerging and serious concern about COVID-19 is the propagation of SARS-CoV-2 RNA mutations16,17,18 in humans or animals, particularly during the development of an effective drug or vaccine against COVID-19.
Following the repeated declarations of emergency in most countries and by the World Health Organization (WHO), the pharmaceutical giants and private and public research organizations in Russia, the United States, the United Kingdom, India, Japan, China, Italy, Spain, Belgium, Germany, Australia, Singapore, and Israel began developing drugs and vaccines against COVID-19 without neglecting the abilities of an RNA virus to bypass an antiviral drug or modify its own genome to override the innate or adaptive immunity of the host. The early episodes of COVID-19 show similarities of SARS-CoV-2 pathogenesis to influenza viruses, HIV (human immunodeficiency virus), Ebola virus, SARS-CoV, and MERS-CoV. Thus, various antiviral drugs have been tested in COVID-19 patients, such as Tamiflu19 and favipiravir20, which are used against common influenza viruses; lopinavir and ritonavir21, which are used against HIV; and remdesivir22, which is used against Ebola virus, Marburg virus, Lass virus, Syncytial virus, Nipah virus, Junin virus, Hendra virus, and CoVs causing SARS and MERS. In addition, several clinical tests were conducted with drugs against malaria-causing Plasmodium falciparum (P. falciparum), such as chloroquine23, mefloquine24, hydroxychloroquine (HCQ)25, artemisinin26, clindamycin27, doxycycline28, and pyrimethamine29. The positive and negative outcomes of these tests alarm tropism switching and genetic modification of the virion. The ongoing treatments of a COVID-19 patient includes the suppression of RNA copying by evading exoribonuclease proofreading, for which the blocking of the endolysosomal transport of the virus-encapsulated endosome is under debate25. In addition, the positive outcome of remdesivir in inhibiting Ebola virus by mutating its RNA is correlated with the positive response of this drug against the first COVID-19 case in the United States30. Nonetheless, the side effects of these drugs, including cardiac malfunction in patients treated with the chloroquine derivatives31, should be carefully considered during the management of COVID-19.
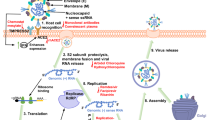
In addition to the aforementioned tests, molecular-level information (Fig. 2) about the infection, immune response, replication, and transmission of SARS-CoV-2 is inevitable during the development of an effective vaccine or a drug. The complexity of the infection and treatment demands a novel platform at the interface of virology, immunology, drug delivery, genetics, chemistry, materials science, and nanoscience, which will help researchers, physicians, and technologists to collaborate and develop vaccines and medicines against COVID-19. The efficient use of the wealth of information about nanomaterials32,33,34,35,36, imaging probes37,38,39,40,41, bioimaging techniques42,43,44,45,46, vaccine development47,48,49,50,51, and in vitro and in vivo drug/gene/nanomaterial delivery52,53,54,55,56,57,58,59,60,61 is essential. Nevertheless, the toxicity of nanomaterials62,63,64,65,66 is a major concern during the consideration of virus mimetic nanoviruses for in vivo applications. This article summarizes the fundamental aspects of the molecular interactions in viral infection and the host immune response and provides future prospects for the aforementioned interface in the fight against the pandemic.
The roles of SGP and ACE2R in viral infection
The membrane-binding proteins of viruses have highly conserved frameworks that are modified according to the available receptors in a host cell. These proteins are β-spirals, similar to those in SGP of SARS-CoV-21, or the coiled-coil α-helix, such as those in HA67 of the influenza virus. We independently consider the roles of SGP-to-ACE2R binding and HA-to-Sia binding (Fig. 2) in SARS-CoV-2 infection. Mechanistically, SARS-CoV-2 infection begins with the recognition of its SGP receptor-binding domain (RBD; Fig. 3) by ACE2R in the host epithelial cells of the respiratory system. The low affinity of ACE2R to its natural ligand is an advantage for the virus. The SGP-to-ACE2R affinity suggests that ACE2Rs in the vascular endothelial cells and myocytes increase the risk and mortality through the systemic transmission of SARS-CoV-2 to vital organs such as the kidneys and the heart68,69. This risk factor is correlated with irregular heartbeats and increased levels of troponin I and creatine kinase in acute COVID-19 patients70. Similarly, physicians are in the early stage of correlating ACE2R and COVID-19 with an immune overreaction and an inflammation in children, which are similar to Kawasaki disease71, affecting vital organs such as the liver, heart, and kidneys.
A A cartoon structure of SARS-CoV-2 showing its SGPs. B The SGP and RBD structures of SARS-CoV-2174. B Copyright of the Royal Society of Chemistry.
The spike in HCQ prescriptions for use against COVID-19 has produced positive and negative results, as well as different opinions about the action of the drug. Although HCQ shows many side effects, including delayed ventricular repolarization and low pulse rates, the success stories of this drug in COVID-19 patients suggest the drug has bimodal action involving the suppression of glycosylation of ACE2R and SGP and the inhibition of RNA release by attenuating the endolysosomal passage of the virion-encompassed endosomes31,72. Thus, HCQ suppresses the release of both SARS-CoV-2 RNA and the progeny virions. Furthermore, HCQ can suppress cytokine production73 and inhibit matrix metalloproteinases74. Nevertheless, the overall outcome for patients treated with HCQ is not positive, which has guided the WHO and many countries to stop recommending this drug for treating COVID-19.
The amino acid residues, particularly five of the six residues critical for SGP-to-ACE2R binding, show a similarity between SARS-CoV-2 and Pangolin-CoV, whereas four of the six critical amino acids in RaTG13 are different7,8,14. However, the presence of a polybasic FCS at the S1–S2 boundary of SGP in SARS-CoV-21,9, which is not common to Pangolin-CoV or other CoVs, increases the affinity of SGP to ACE2R. The deep canyon formed at the S1–S2 junction of SGP enables the efficient binding of the virus to ACE2R. Following binding, the proteolytic S1–S2-cleaved subunits endow SGP with a 10- to 20-fold greater affinity for ACE2R and a more infectious nature to SARS-CoV-2 than SARS-CoV. In contrast, the trimeric RBD of SARS-CoV is conserved throughout the infection period. The S1 subunit helps SARS-CoV-2 bind to ACE2R, and S2 enables the entry of the virus into a host cell. The free-energy change accompanying an SGP-induced conformational change to an ACE2R increases the infectivity by favoring the binding of another SGP of the same or different virus to a proximal receptor. Indeed, the energy barrier for appropriately destabilizing the host cell membrane and membrane fusion is as high as 42 kcal/mol, which can be supplemented by the low-affinity pseudo or secondary receptors in a host cell, such as heparin sulfate, ceramide derivatives, and HA.
The genes for FCS in SARS-CoV-2 suggest that a mutation occurred in R. affinis, M. javanica, or humans. Nevertheless, S1–S2 cleavage and the specificity of SGP to ACE2R indicate repeated passages of the SARS-CoV-2 progenitor in host cells expressing ACE2R, such as in humans, pigs, or civets. Thus, we do not rule out the possibility that the progenitor infected a human and repeatedly passed through one or more persons, who may or may not have shown COVID-19 symptoms. To date, there is no evidence to suggest that SARS-CoV mutated to SARS-CoV-2 in a cell or an animal model. Nevertheless, the mutations to SARS-CoV-2 continue to emerge in COVID-19 patients and is likely to continue further through animal and human hosts because, similar to other CoVs, SARS-CoV-2 virus lacks an enzyme for RNA proofreading, and preparations are necessary to deal with more strains that are more aggressively infectious than SARS-CoV-2. Despite the involvement of FCS, SGP, and ACE2R moieties in infection, the roles of Sias and HAs in SARS-CoV-2 and coreceptors such as chemokine receptors and O-linked glycan receptors in host cells should be considered during not only the development of a vaccine or a drug but also selecting foods and supplements to fight COVID-19.
The roles of Sia and HA in the infection and replication
Sia-capped proteoglycans in cells, secreted molecules, and vaccines play key roles in cell repulsion/adhesion, embryogenesis, pathogenesis, and the immune response75,76. HA (Fig. 4A), a lectin with ligand-binding domains 1 and 2, is common to CoVs. The binding of a pathogen to a Sia, such as the HA-based binding of CoVs to N-acetylneuraminic acid (Neu5Ac) (Fig. 4B), is the first step of an infection77,78. Thus, HA-to-Sia binding is addressed in several experimental and theoretical studies involving HA of influenza viruses77,79,80,81. Domain 1 (Fig. 4C) of HA shows a high affinity for Sia-capped glycans in a host cell77. Similarly to HA enrichment in an inactivated influenza vaccine, we hypothesize that one of the aims of COVID-19 management can be the HA-based binding of SARS-CoV-2 to proteoglycans in a host cell. We suggest this strategy by considering the binding of a mutated HA in the avian influenza virus to Neu5Ac in human epithelial cells75,77,82. Neu5Ac is human-specific α2–6-linked Sia that caps the outermost parts of proteoglycans in cell membranes and secretions. Sia derivatives inhibit HA and prevent the attachment of influenza viruses to cells77. In contrast, the native HA of a CoV selectively binds to N-glycolylneuraminic acid (Neu5Gc) in a bird and the bird-to-human transmission is inefficient83. While several studies have focused on mutations to SGP and SGP-to-ACE2R binding, less attention has been directed to the roles of HA and Neu5Ac in SARS-CoV-2 and the levels of Neu5Ac in the upper respiratory system and the lungs of COVID-19 patients. Nonetheless, the affinity of HA for Neu5Gc should be considered. The presence of HAs and Neu5Ac-selective NAs in CoVs directed our attention to the Sia-bearing glycans, and the pathological and immunological aspects of COVID-19.
The structures of A HA, B the Neu5Ac segment of Sia, and C the Sia-binding domain of HA. Courtesy of D. Sehnal, A. S. Rose, J. Kovca, S. K. Burley, S. Velankar (2018), RCSB Protein Data Bank 3GBN.
Similarly to other pathogens, CoVs express Sia-bearing glycans on their surfaces to deceive sensors that trigger IFN signaling and bypass host innate immunity. Thus, vertebrates modify their own glycans to fight pathogens. To comprehend the HA- and Neu5Ac-based infection of the respiratory epithelial cells by SARS-CoV-2, we compare the structures (Fig. 4) of Neu5Ac and the Sia-binding domain of the HA peptide. The multiple hydroxyl moieties in Neu5Ac, which are hydrogen (H)-bond donors, and the multiple carboxylic and amide moieties in the HA domain, which are H-bond acceptors, can be the thermodynamic regulators of the binding and infection. The strength of the H-bonds formed between HA and Neu5Ac is assessed based on the free-energy change of biotin–streptavidin (B-S) complex formation (Fig. 5A)84. Although the net entropy change (−13.7 kcal/mol) does not favor the formation of the B-S) complex, the enthalpic contribution (−32 kcal/mol) stabilizes it84,85,86. In the B-S complex, the enthalpy change involves multiple H-bonds of the ureido and carboxylic groups in biotin to seven amino acids in avidin. Similarly, by considering multiple H-bond donors and acceptors in the HA domain and Neu5Ac, we calculate the H-bond energy of the HA–Neu5Ac complex at 34.3 kcal/mol using the B3LYP/6–31 + G** level of the density functional theory87. The initial structures of the HA domain and Neu5Ac were obtained from the Protein Data Bank88. In addition, the initial relative positions of the two were assumed from the structures in the data bank. The heat of HA-to-Neu5Ac complexation is not surprising to us by considering the eight H-bonds between α2-3-linked sialyllactose and the eight amino acids in the Sia-binding domain of HA77. The appreciably high enthalpy of H-bonding stabilizes the HA–Neu5Ac complex more than it does in the B-S complex, enabling the HA–Neu5Ac complex to form deep potential well that attracts a virus and a progeny virion, which benefit from energy-efficient NA/ES-hydrolyzed cleavage during endocytosis and release.
Although the HA-mediated binding of pathogens to Neu5Ac in epithelial cells is common to many diseases, an important lineage difference between chimpanzees and humans, that is, the presence of Ne5Gc in place of Ne5Ac in chimpanzees, is neglected during clinical tests of the antimalarial drugs such as HCQ against SARS-CoV-2. While chimpanzees or bonobos injected with P. falciparum-infected blood or exposed to P. falciparum-carrying Anopheles mosquitoes do not show the malaria symptoms, P. falciparum infects human erythrocytes expressing Neu5Ac and causes life-threatening malaria89,90,91,92. In contrast, Plasmodium reichenowi, which shares a common ancestor with P. falciparum, effectively infects apes expressing Neu5Gc in their cell membrane93. Thus, in addition to the FCS modification and ACE2R-based binding of SARS-CoV-2 onto human respiratory epithelial cells, lineage-derived Neu5Ac capping of epithelial glycoproteins may be at the center of the human-selective lethal infection of SARS-CoV-2 through HA. Although HCQ suppresses the endosomal escape of SARS-CoV-2, it does not show any relation to Neu5Ac in humans or Neu5Gc in chimpanzees. Nevertheless, the risk factors for COVID-19 may be related to the presence of any Neu5Gc-specific agglutinin in SARS-CoV-2 and a high level of Neu5Gc-capped glycans acquired through the assimilation of animal proteins by the COVID-19 patient. This HA- and Sia-based pathogenesis becomes significant to CoVs bearing an ES and a NASe specific to the α2-3-linked Sias. Although the T and B cells with the α2-6-linked glycans and the macrophages surveilling for the Neu5Ac-bearing pathogens play crucial roles in the immunosuppression of HA-to-Sia selective infections, we suggest that verifying the relationships of the acquired Neu5Gc to various agglutinins, O-linked glycans, and ESes and NASes is important. This verification can be relevant to COVID-19 because of the different infection and mortality rates of the disease in populations with different dietary habits, and lineages.
Pattern recognition, immune response, IFN production, and virus replication
An immune response against SARS-CoV-2, similarly to that against other CoVs and microbes, begins with the activation of a series of pattern recognition receptors (PRRs). During surface binding, internalization, endosomal transport, cytosolic uncoating, RNA polymerization, and translation in a host cell, viral lipids, proteins, and RNA are recognized by various pathogen-associated molecular patterns (PAMPs) in the cell membrane, endosomes, and cytoplasm. This recognition continues throughout the viral lifecycle in a host cell. Figure 6 shows various PAMP- and PRR-associated immune responses in COVID-19, which are analogous to other CoVs. In contrast to a nonenveloped virus that enters a host cell by the endocytic/nonendocytic path94,95, the enveloped nature of SARS-CoV-2 leads to its preferential endocytic membrane fusion through the recruitment of clathrin/calveolin96,97. The mechanisms of infection involving the virological synapse, cell penetration, and transcytosis have yet to be verified. SARS-CoV-2 binds to a host cell through the SGP-TMPSS2-ACE2R (SGP-transmembrane protease serine 2-ACE2R) network1,8,14,68,69. In SGP-to-ACE2R binding, the free-energy change of the conformation switching of a single SGP enables membrane fusion98.
Following the extracellular ACE2R-specific binding using SGP, and the different stages of pattern recognition, the immune responses in COVID-19 are classified according to the activation of PAMPs by various viral patterns (Fig. 6). In general, pattern recognition is initiated by the interactions of surface proteins, genetic materials (single-stranded RNA (ssRNA)/double-stranded DNA (dsRNA)/ssDNA/dsDNA), or uncoated/translated proteins of a pathogen by PRRs, such TLRs (toll-like receptors)99,100,101,102,103, RLRs (RIG-like receptors)104,105,106,107, NLRs (NOD-like receptors)108,109,110,111,112, MDA-5 (melanoma differentiation-associated protein 5)113,114, CLR/MR (C-type lectin-like receptor/mannose receptor)115,116, and DAI (DNA-dependent activator of IFN-regulatory factors (IRFs))117,118 in a host cell. For example, the dsRNA of the nonenveloped Reoviridae family of viruses, such as rotavirus, is recognized by TLR3, RIG-I (retinoic acid-inducible gene I), and MDA-5119,120,121, whereas the ssRNA of viruses such as SARS-CoV-2, SARS-CoV, MERS-CoV, rhinoviruses, dengue virus, and hepacivirus C are recognized in the endosome by TLR7 and TLR8122,123,124,125,126,127. In contrast, the ssDNA of viruses such as the smallpox virus and the chickenpox or varicella viruses is effectively recognized by TLR9, and RIG-I128,129. Indeed, independent of the DNA genome of various viruses, pattern recognition, and IFN induction follow the RNA pathway because of reverse transcription during replication.
Various PRRs and PAMPs involved in SARS-CoV-2 and other pathogens are summarized in Fig. 6 and briefly discussed here. PAMP1 is initiated by the recognition of viral lipopolysaccharide (LPS) by TLR4130 followed by its association with CD14, leading to the activation of the MyD88-dependent131 and MyD88-independent132 pathways and the secretion of several cytokines, including interleukin-1β (IL-1β)/6/8/12, and IFN-α/β/γ. Extracellular PAMP2 is recognized by TLR4133,134, which associates with CLRs such as MR and activates the complex MAPK downstream signaling. Only a part of the MAPK pathway leading to the secretion of IL-1β/2/6/10/18, and TNF-α is shown in Fig. 6. The cytosolic ion imbalance forms PAMP3135,136, which is created by ion channel mimicking nonstructural viral proteins137,138, pathogen-associated Ca2+ influx, or K+ efflux. PAMP4 is triggered by ssRNA139,140 released from the endosomes, subgenomic RNAs produced by the RNA-dependent RNA polymerase, or the RNA fragments produced by RNASe-L. PAMP3 and PAMP4 are recognized by NLRP3135,136,137,138,139,140,141, leading to the activation of multiple signaling pathways by Casp-I and the secretion of various ILs, and TNF-α, which are shown in Fig. 6. The NLRP3 inflammasome pathways are excluded here. PAMP5 is a combination of viral proteins, including those uncoated and released from endosomes, and those translated by reading the genomic/subgenomic viral RNA. PAMP5 is recognized by RLR142,143,144, which associates with MDA-5 and MAVS (mitochondrial antiviral signaling) and activates IRF3/7, leading to the secretion of IFN-α/β. PAMP6 is the endosomal recognition of ssRNA by TLR7/8124,125,145,146,147, leading to multiple signaling pathways through TNF receptor-associated factors (IκB kinases and p38), and the secretion of IL-1/6, IFN-α/β, and TNF-α.
IFNs148,149,150,151,152,153,154,155,156,157, a family of immunomodulatory macromolecular ligands or secreted cytokines play an important role in an immune response against SARS-CoV-2. Apart from the important antitumor and antiproliferative roles of IFNs158,159, these cytokines help fight viral infections by activating innate and adaptive immune responses. SARS-CoV-2 pattern-recognized secretion of IFNs is shown in Fig. 6. On the one hand, pattern recognition followed by downstream signaling pathways produces ILs, IFNs, and TNF-α and on the other hand, antiviral action is triggered in the JAK-STAT transcription pathway, which is by secondary signaling and the activation of IFN receptors in the infected and immune cells148,149. Among the 23 widely known members in the IFN family in mammals, 21 are common to the human body. Based on functioning, signaling pathways, and clustering in chromosomes, IFNs are classified into IFN-I, -II, and -III. IFN-I is the largest family, with 19 members for which the genes are clustered on human chromosome 9150. Among the 19 members, IFN-α (13 homologs) and IFN-β distributed in essentially all cells play the primary role in the innate response against infections, through the JAK-STAT pathway. IFN-II, also called IFN-γ, with its genes clustered in human chromosome 12151, is common to immune cells, and its secretion and action are delayed until the immune cells are activated. Thus, IFN-γ is the adaptive cytokine. IFN-III includes three members, IFN-λ1, -λ2, and -λ3, also called ILs 28A, 28B, and 29, with their genes clustered in human chromosome 19152. Similar to IFN-I cytokines, IFN-III cytokines are induced by viruses and are secreted by any cell in response to SARS-CoV-2. Thus, IFN-I and IFN-III secreted by the respiratory epithelial cells initiate innate immunomodulation against SARS-CoV-2. The JAK-STAT pathway of secondary signaling is discussed elsewhere148,149.
Following asymptomatic infection by SARS-CoV-2, a person with a weak immune system becomes sick and suffers from a severe immune overreaction or cytokine storm, leading to an increased respirational distress and fatality153. Thus, immune suppressants such as Actemra or tocilizumab are prescribed to COVID-19 patients159. Nonetheless, the recovery rate of COVID-19 patients depends on the health condition of the patients. For example, patients younger than 40 years old with diabetes, cardiovascular diseases, impaired renal health, or cancer show a poor recovery rate after COVID-19 infection160,161. The relationship between the innate and adaptive immune (IFN) response of host cells, including the production of various proinflammatory cytokines, IFN-stimulated genes, IRFs, and various proteins including myxovirus (Mx) proteins and the ability of the virus to escape the immune response are important to the recovery of a COVID-19 patient. Mutations to the RNA of SARS-CoV-2 when compared with those in Pangolin-CoV, MERS-CoV, SARS-CoV, and RaTG13 may provide clues about turning off or overstimulating the IFN pathway in COVID-19. The cytokine storm in several fatal COVID-19 cases suggests that the virus does not bypass pattern recognition (Fig. 6). Thus, the role of IFN-inducible genes such as ISGs and Mx-A/-B proteins, which are crucial for preventing the formation of progeny virions through both the inactivation of the nucleocapsid (NC) protein and inhibition of the progeny ribonucleoprotein, in the cytokine storm and the progression of COVID-19 is being extensively investigated.
To date, there is no strong experimental correlation between SARS-CoV-2 and any acute respiratory illness in mice or pets. In addition, in contrast to humans, several animals do not show functional IFNs. Thus, the long lineage from a common ancestor of humans and animals should be the basis for a search for a correlation between IFN-inducible Mx genes and COVID-19. In addition to the presence of Mx-inducible genes in humans, the defective Mx genes in mice susceptible to influenza162 and the IFN-regulated Mx in mice resistant to influenza163 educate us on the importance of examining how SARS-CoV-2 inactivates the innate IFN-I/III pathways and successfully integrates progeny virions.
Summary and outlook
Despite the auspicious progress of COVID-19 vaccine research and vaccination, the highly infectious nature and mutations of SARS-CoV-2 are warnings of a backbiting annual revival of the virus. Evidence from RNA-sequencing showing the infection mode of SARS-CoV-2 to be SGP-to-ACE2R binding is supported by cleavage at the S1–S2 boundary of the furin segment. On the one hand, ACE2R in the respiratory endothelial cells is involved in the infection, and on the other hand, its presence in the cells of the heart, kidneys, and other vascular systems underscores the relationship of COVID-19 to cardiovascular diseases, cancer, and diabetes. Also, ACE2R-based infection may be related to the Kawasaki disease-mimetic multisystem inflammatory syndrome in children. In addition to ACE2R-based binding, the roles of HA, Neu5Ac(Ge), and NASe/SE in SARS-CoV-2 infection and release cannot be neglected.
The relationship between Sias and HA has attracted much attention since the 1918 influenza (H1N1) pandemic, whereas the details of the binding between the two are the focus of ongoing studies. Enzymes such as NASe, which cleaves the bond between the viral HA and the host Sias164, likely have roles in the endocytosis of SARS-CoV-2 in the respiratory epithelial cells and the release of its progeny through the ER pathway. Although the Neu5Ac Sia innate to humans is the target of several pathogens, including CoVs, the negative results for COVID-19 patients treated with several antiviral drugs and vaccines may be related to mutations in not only the FCS motif but also the Neu5Ac-binding segment of HA. If a innate Sia is not the target of the SARS-CoV-2 HA, it is worth considering a connection between the levels of Sias acquired from the diet, such as Neu5Gc, and the severity of COVID-19. Another aspect of Sias in COVID-19 is the escape of the virus from the innate IFN response, which is similarly to other pathogens that express and modify Sias on the cell surface. In addition, the abilities of Sia derivatives to inhibit HA and thwart influenza viruses encourage us to hypothesize that dietary supplements with acidic sugars, such as ascorbic acid, gluconic acid, saccharic acid, and tartaric acid, may have impacts on COVID-19. Although clinical studies are needed to understand whether Neu5Ac, Neu5Gc, other Sias, or sugar acids alleviate the severity of COVID-19, diets rich in sugar acid-rich fruits can always be recommended to a COVID-19 patient. Nevertheless, the stereoselectivity of sugar acids, Sias, and NASe/SE needs further study.
To resolve the preclinical challenges of COVID-19, an interface among chemistry, nanoscience, cell biology, and virology is emerging165,166,167,168,169,170,171,172,173. The primary objective can be the construction of a SARS-CoV-2 mimetic virus nanoparticle (SCoV-MNP; Fig. 7) decorated with SPG, HA, and Sias and encompassed by ESs/NASes but without the SARS-CoV-2 RNA. Subsequently, the microspectroscopic correlation of SPG- or HA-based binding of SCoV-MNPs to Sias in human respiratory/lung epithelial cells has helped increase the understanding of the mode of infection. In addition, SCoV-MNPs decorated with proton-buffering molecules will be helpful for revealing the endolysosomal pathway and the viral uncoating processes. Such virus mimetics can be modified into antiviral drugs engaged in the endosomal arrest of SARS-CoV-2. Fluorescence-assisted cell sorting, confocal fluorescence microscopy, super-resolution fluorescence imaging, and Förster resonance energy transfer-microspectroscopy are powerful tools for analyzing the binding, membrane fusion, endocytosis, uncoating, the formation of progeny virions, and the release of SARS-CoV-2. In such studies, fluorescence probes and energy donors can be selected from among organic dyes, fluorescent proteins, or brilliantly luminescent semiconductor quantum dots. These studies will be helpful for evaluating the stereoselectivity of ESs and NASes in SCoV-MNPs to O-linked glycans. In addition, the escape of CoVs from the host innate immune response can be analyzed using SCoV-MNPs decorated with Sias.
References
Wrapp, D. et al. Cryo-EM structure of the 2019-NCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Goldsmith, C. S., Miller, S. E., Martines, R. B., Bullock, H. A. & Zaki, S. R. Electron microscopy of SARS-CoV-2: a challenging task. Lancet 395, e99 (2020).
To, K. K. W. et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 20, 565–574 (2020).
Lu, R. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 (2020).
Jeffers, S. A. et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl Acad. Sci. USA 101, 15748–15753 (2004).
Agostini, M. L. et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio 9, 1–15 (2018).
Zhang, T., Wu, Q. & Zhang, Z. Probable Pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 30, 1346–1351 (2020).
Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C. & Garry, R. F. The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020).
Angeletti, S. et al. COVID-2019: the role of the Nsp2 and Nsp3 in its pathogenesis. J. Med. Virol. 92, 584–588 (2020).
Liu, Z. et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 92, 595–601 (2020).
Bouhaddou, M. et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182, 685–712 (2020).
Fensterl, V. & Sen, G. C. Interferons and viral infections. Biofactors 35, 14–20 (2009).
Haller, O. & Kochs, G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interf. Cytokine Res. 31, 79–87 (2011).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Guo, Y. R. et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil. Med. Res. 7, 110 (2020).
Letko, M., Marzi, A. & Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569 (2020).
Zhang, X. Epidemiology of Covid-19. N. Engl. J. Med. 382, 1869–1869 (2020).
Ali Hosseini Rad, S. M. & McLellan, A. D. Implications of Sars-Cov-2 mutations for genomic RNA structure and host microRNA targeting. Int. J. Mol. Sci. 21, 1–18 (2020).
Sanders, J. M., Monogue, M. L., Jodlowski, T. Z. & Cutrell, J. B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 323, 1824–1836 (2020).
Tu, Y. et al. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 21, 2657 (2020).
Chong, V. H. et al. Conduction abnormalities in hydroxychloroquine add on therapy to lopinavir/ritonavir in COVID-19. J. Med. Virol. 92, 1–3 (2020).
Durante-Mangoni, E. et al. Early experience with remdesivir in SARS-CoV-2 pneumonia. Infection 48, 779–782 (2020).
Wang, M. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-NCoV) in vitro. Cell Res. 30, 269–271 (2020).
Fan, H. H. et al. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chin. Med. J. 133, 1051–1056 (2020).
Colson, P., Rolain, J. M., Lagier, J. C., Brouqui, P. & Raoult, D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents 55, 105932 (2020).
Sehailia, M. & Chemat, S. Antimalarial-agent artemisinin and derivatives portray more potent binding to Lys353 and Lys31-binding hotspots of SARS-CoV-2 Spike protein than hydroxychloroquine: potential repurposing of artenimol for COVID-19. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2020.1796809 (2020).
Tan, S. H. S., Hong, C. C., Saha, S., Murphy, D. & Hui, J. H. Medications in COVID-19 patients: summarizing the current literature from an orthopaedic perspective. Int. Orthop. 44, 1599–1603 (2020).
Sodhi, M. & Etminan, M. Therapeutic potential for tetracyclines in the treatment of COVID-19. Pharmacotherapy 40, 487–488 (2020).
Kuupiel, D. et al. Geographical accessibility to glucose-6-phosphate dioxygenase deficiency point-of-care testing for antenatal care in Ghana. Diagnostics 10, 1–14 (2020).
Holshue, M. L. et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382, 929–936 (2020).
Meyerowitz, E. A. et al. Rethinking the role of hydroxychloroquine in the treatment of COVID-19. FASEB J. 34, 6027–6037 (2020).
Chouhan, L., Ghimire, S., Subrahmanyam, C., Miyasaka, T. & Biju, V. Synthesis, optoelectronic properties and applications of halide perovskites. Chem. Soc. Rev. 49, 2869 (2020).
Biju, V., Itoh, T., Anas, A., Sujith, A. & Ishikawa, M. Semiconductor quantum dots and metal nanoparticles: syntheses, optical properties, and biological applications. Anal. Bioanal. Chem. 391, 2469 (2008).
Biju, V., Itoh, T. & Ishikawa, M. Delivering quantum dots to cells: bioconjugated quantum dots for targeted and nonspecific extracellular and intracellular imaging. Chem. Soc. Rev. 39, 3031 (2010).
Usman, K. A. S. et al. Downsizing metal–organic frameworks by bottom-up and topdown methods. NPG Asia Mater. 12, https://doi.org/10.1038/s41427-020-00240-5 (2020).
Murray, C. B., Norris, D. J. & Bawendi, M. G. Synthesis and characterization of nearly monodisperse CdE (E = S, Se, Te) semiconductor nanocrystallites. J. Am. Chem. Soc. 115, 8706 (1993).
Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 43, 744 (2014).
Biju, V. et al. Nanoparticles speckled by ready-to-conjugate lanthanide complexes for multimodal imaging. Nanoscale 7, 14829 (2015).
Biju, V. et al. Quantum dot-insect neuropeptide conjugates for fluorescence imaging, transfection, and nucleus targeting of living cells. Langmuir 23, 10254 (2007).
Liong, M. et al. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS Nano 2, 889 (2008).
Dykman, L. & Khlebtsov, N. Gold nanoparticles in biomedical applications: recent advances and perspectives. Chem. Soc. Rev. 41, 2256 (2012).
Shibu, E. S. et al. Singlet-oxygen-sensitizing near-infrared-fluorescent multimodal nanoparticles. Angew. Chem. Int. Ed. 52, 10559 (2013).
Hell, S. W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780 (1994).
Kawashima, N., Nakayama, K., Itoh, K., Ishikawa, M. & Biju, V. Reversible dimerization of EGFR revealed by single-molecule fluorescence imaging using quantum dots. Chem. Eur. J. 16, 1186 (2010).
Huang, X., El-Sayed, I. H., Qian, W. & El-Sayed, M. A. Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. J. Am. Chem. Soc. 128, 2115 (2006).
Qian, X. et al. In vivo tumor targeting and spectroscopic detection with surface enhanced raman nanoparticle tags. Nat. Biotechnol. 26, 83 (2008).
Prompetchara, E., Ketloy, C. & Palaga, T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 38, 1 (2020).
Ahmed, S. F., Quadeer, A. A. & McKay, M. R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 12, https://doi.org/10.3390/v12030254 (2020).
Gurwitz, D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 81, 537 (2020).
Tai, W. et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 17, 613 (2020).
Liu, C. et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 6, 315 (2020).
Biju, V. et al. FRET from quantum dots to photodecompose undesired acceptors and report the condensation and decondensation of plasmid DNA. ACS Nano 6, 3776 (2012).
Anas, A. et al. Clathrin-mediated endocytosis of quantum dot-peptide conjugates in living cells. ACS Nano 3, 2419 (2009).
Biju, V., Mundayoor, S., Omkumar, R. V., Anas, A. & Ishikawa, M. Bioconjugated quantum dots for cancer research: present status, prospects and remaining issues. Biotechnol. Adv. 28, 199 (2010).
Shibu, E. S. et al. Photouncaging nanoparticles for MRI and fluorescence imaging in vitro and in vivo. ACS Nano 7, 9851 (2013).
Yuan, Z. et al. Novel fluorinated polycationic delivery of anti-VEGF siRNA for tumor therapy. NPG Asia Mater. 12, https://doi.org/10.1038/s41427-020-0216-9 (2020).
Mu, L. M. et al. Development of functional dendrisomes based on a single molecule of polyesterbenzylether dendrimer and their application in cancer stem cell therapy. NPG Asia Mater. 11, https://doi.org/10.1038/s41427-019-0134-x (2019).
Tunesi, M. et al. Hydrogel-based delivery of Tat-fused protein Hsp70 protects dopaminergic cells in vitro and in a mouse model of Parkinson’s disease. NPG Asia Mater. 11, https://doi.org/10.1038/s41427-019-0128-8(2019).
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358 (2019).
Cai, W. et al. Metal–organic framework-based stimuli-responsive systems for drug delivery. Adv. Sci. 6, https://doi.org/10.1002/advs.201801526 (2019).
Kauscher, U., Holme, M. N., Björnmalm, M. & Stevens, M. M. Physical stimuli responsive vesicles in drug delivery: beyond liposomes and polymersomes. Adv. Drug Deliv. Rev. 138, 259 (2019).
Jones, P., Sugino, S., Yamamura, S., Lacy, F. & Biju, V. Impairments of cells and genomic DNA by environmentally transformed engineered nanomaterials. Nanoscale 5, 9511 (2013).
Sobhanan, J. et al. Toxicity of nanomaterials due to photochemical degradation and the release of heavy metal ions. Nanoscale. https://doi.org/10.1039/d0nr03957h (2020).
Derfus, A. M., Chan, W. C. W. & Bhatia, S. N. Probing the cytotoxicity of semiconductor quantum dots. Nano Lett. 4, 11 (2004).
Lewinski, N., Colvin, V. & Drezek, R. Cytotoxicity of nanopartides. Small 4, 26 (2008).
De Jong, W. H. & Borm, P. J. A. Drug delivery and nanoparticles: applications and hazards. Int. J. Nanomed. 3, 133 (2008).
Carr, C. M. & Kim, P. S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73, 823–832 (1993).
Gallagher, P. E., Ferrario, C. M. & Tallant, E. A. Regulation of ACE2 in cardiac myocytes and fibroblasts. Am. J. Physiol. Heart Circ. Physiol. 295, 2373–2379 (2008).
Mendoza Torres, E. et al. ACE2 and vasoactive peptides: novel players in cardiovascular/renal remodeling and hypertension. Ther. Adv. Cardiovasc. Dis. 9, 217–237 (2015).
Zhou, F. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395, 1054–1062 (2020).
Newburger, J. W. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a Statement for Health Professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young. Am. Heart Assoc. Circ. 110, 2747–2771 (2004).
Mourad, J. J. & Levy, B. I. Interaction between RAAS Inhibitors and ACE2 in the context of COVID-19. Nat. Rev. Cardiol. 17, 313 (2020).
Zhao, M. Cytokine storm and immunomodulatory therapy in COVID-19: role of chloroquine and anti-IL-6 monoclonal antibodies. Int. J. Antimicrob. Agents 55, 105982 (2020).
Xu, F. et al. Development of a PH-responsive polymersome inducing endoplasmic reticulum stress and autophagy blockade. Sci. Adv. 6, eabb8725 (2020).
Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 14, 351–360 (2008).
Paulson, J. C. 5—Interactions of animal viruses with cell surface receptors. Receptors 2, 131–219 (1985).
Sauter, N. K. et al. Binding of influenza virus hemagglutinin to analogs of its cellsurface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry 31, 9609–9621 (1992).
Wasik, B. R., Barnard, K. N. & Parrish, C. R. Effects of sialic acid modifications on virus binding and infection. Trends Microbiol. 24, 991–1001 (2016).
Wiley, D. C. & Skehel, J. J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56, 365–394 (1987).
Carroll, S. M., Higa, H. H. & Paulson, J. C. Different cell-surface receptor determinants of antigenically similar influenza virus hemagglutinins. J. Biol. Chem. 256, 8357–8363 (1981).
Higa, H. H., Rogers, G. N. & Paulson, J. C. Influenza virus hemagglutinins differentiate between receptor determinants bearing N-acetyl-, N-glycollyl-, and N, Odiacetyineuraminic acids. Virology 144, 279–282 (1985).
Gagneux, P. et al. Human-specific regulation of Α2-6-linked sialic acids. J. Biol. Chem. 278, 48245–48250 (2003).
Chen, Y. et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381, 1916–1925 (2013).
Weber, P. C., Wendoloski, J. J., Pantoliano, M. W. & Salemme, F. R. Crystallographic and thermodynamic comparison of natural and synthetic ligands bound to streptavidin. J. Am. Chem. Soc. 114, 3197–3200 (1992).
Stayton, P. S. et al. Streptavidin-biotin binding energetics. Biomol. Eng. 16, 39–44 (1999).
Chivers, C. E., Koner, A. L., Lowe, E. D. & Howarth, M. How the biotin-streptavidin interaction was made even stronger: investigation via crystallography and a chimaeric tetramer. Biochem. J. 435, 55–63 (2011).
Chen, J. et al. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95, 409–417 (1998).
Kruse, H., Goerigk, L. & Grimme, S. Why the standard B3LYP/6-31G* model chemistry should not be used in DFT calculations of molecular thermochemistry: understanding and correcting the problem. J. Org. Chem. 77, 10824–10834 (2012).
Blacklock, B. & Adler, S. A parasite resembling Plasmodium falciparum in a chimpanzee. Ann. Trop. Med. Parasitol. 160, 99–106 (1992).
Rodhain, J. Les plasmodium des anthropoids de l’Afrique centrale et leur relation avec les plasmodium humains. Ann. Soc. Belg. Med. Trop. 19, 563–572 (1939).
Ollomo, B. et al. Lack of malaria parasite transmission between apes and humans in Gabon. Am. J. Trop. Med. Hyg. 56, 440–445 (1997).
Martin, M. J., Rayner, J. C., Gagneux, P., Barnwell, J. W. & Varki, A. Evolution of human–chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl Acad. Sci. USA 102, 12819–12824 (2005).
Escalante, A. A. & Ayala, F. J. Phylogeny of the malarial genus plasmodium, derived from RRNA gene sequences. Proc. Natl Acad. Sci. USA 91, 11373–11377 (1994).
Kalia, M. & Jameel, S. Virus entry paradigms. Amino Acids 41, 1147–1157 (2011).
Klasse, P. J., Bron, R. & Marsh, M. Mechanisms of enveloped virus entry into animal. Cells Adv. Drug Deliv. Rev. 34, 65–91 (1998).
Glebov, O. O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 287, 3664–3671 (2020).
Celik, O. et al. Combating sars-cov-2 through lipoxins, proteasome, caveolin and nuclear factor-kappa b pathways in non-pregnant and pregnant populations. Cell. Mol. Biol. 66, 221–229 (2020).
Gur, M. et al. Conformational transition of SARS-CoV-2 spike glycoprotein between its closed and open states. J. Chem. Phys. 153, 075101 (2020).
Akira, S., Takeda, K. & Kaisho, T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680 (2001).
Akira, S. & Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 (2004).
Beutler, B. Inferences, questions and possibilities in Toll-like receptor signaling. Nature 430, 257–263 (2004).
Trinchieri, G. & Sher, A. Cooperation of Toll-like receptor signals in innate immune defense. Nat. Rev. Immunol. 7, 179–190 (2007).
Barton, G. M. & Medzhitov, R. Toll-like receptor signaling pathways. Science 300, 1524–1525 (2003).
Kowalinski, E. et al. Structural basis for the activation of innate immune patternre cognition receptor Rig-I by viral RNA. Cell 147, 423–435 (2011).
Loo, Y. M. & Gale, M. Jr Immune signaling by RIG-I-like receptors. Immunity 34, 680–692 (2011).
Yoneyama, M. & Fujita, T. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 282, 15315–15318 (2007).
Rehwinkel, J. & Gack, M. U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 20, 537–551 (2020).
Kanneganti, T. D., Lamkanfi, M. & Nunez, G. Intracellular NOD-like receptors in host defense and disease. Immunity 27, 549–559 (2007).
Wen, H. T., Miao, E. A. & Ting, J. P.-Y. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity 39, 432–441 (2013).
Geddes, K., Magalhaes, J. G. & Girardin, S. E. Unleashing the therapeutic potential of Nod-like receptors. Nat. Rev. Drug Discov. 8, 465–479 (2009).
Eisenbarth, S. C. et al. NLRP10 is a NOD-like receptor essential to initiate adaptive immunity by dendritic cells. Nature 484, 510–513 (2012).
Feerick, C. L. & McKernan, D. P. Understanding the regulation of pattern recognition receptors in inflammatory diseases—a ‘NOD’ in the right direction. Immunology 150, 237–247 (2017).
Wu, B. et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152, 276–289 (2013).
Kato, H. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006).
Geijtenbeek, T. B. H. & Gringhuis, S. I. Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 9, 465–479 (2009).
Stahl, P. D. & Ezekowitz, R. A. B. The mannose receptor is a pattern recognition receptor involved in host defense. Curr. Opin. Immunol. 10, 50–55 (1998).
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–504 (2007).
Kuriakose, T. et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 1, aag2045 (2016).
Matsumoto, M. et al. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J. Immunol. 171, 3154–162 (2003).
Broquet, A. H., Hirata, Y., McAllister, C. S. & Kagnoff, M. F. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 186, 1618–1626 (2011).
Sen, A., Pruijssers, A. J., Dermody, T. S., Garcia-Sastre, A. & Greenberg, H. B. The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF3. J. Virol. 85, 3717–3732 (2011).
Sallenave, J.-M. & Guillot, L. Innate immune signaling and proteolytic pathways in the resolution or exacerbation of SARS-CoV-2 in Covid-19: key therapeutic targets? Front. Immunol. 11, 1229 (2020).
Choudhury, A. & Mukherjee, S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. https://doi.org/10.1002/jmv.25987 (2020).
Heil, F. et al. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303, 1526–1529 (2004).
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S. & Sousa, C. R. E. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004).
Yang, K. et al. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and lambda is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity 23, 465–478 (2005).
Lund, J. M. et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl Acad. Sci. USA 101, 5598–5603 (2004).
Harte, M. T. et al. The Poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 197, 343–351 (2004).
Lester, S. N. & Li, K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 426, 1246–1264 (2014).
Kurt-Jones, E. A. et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1, 398–401 (2000).
Fitzgerald, K. A. et al. Mal (MyD88-Adapter-Like) is required for Toll-like receptor4 signal transduction. Nature 413, 78–83 (2001).
Yamamoto, M. et al. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301, 640–643 (2003).
Gronwall, C. et al. MAPK phosphatase-1 is required for regulatory natural autoantibody-mediated inhibition of TLR responses. Proc. Natl Acad. Sci. USA 109, 19745–19750 (2012).
Ip, W. K. E., Takahashi, K., Ezekowitz, R. A. & Stuart, L. M. Mannose-binding lectin and innate immunity. Immunol. Rev. 230, 9–21 (2009).
Kawai, T. & Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21, 317–337 (2009).
Katsnelson, M. A., Lozada-Soto, K. M., Russo, H. M., Miller, B. A. & Dubyak, G. R. NLRP3 inflammasome signaling is activated by low-level lysosome disruption but inhibited by extensive lysosome disruption: roles for K+ efflux and Ca2+ influx. Am. J. Physiol. Cell Physiol. 311, C83–C100 (2016).
Ichinohe, T., Pang, I. K. & Iwasaki, A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 11, 404–410 (2010).
Alexander, S. P. H. et al. Rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development. Br. J. Pharmacol. https://doi.org/10.1111/bph.15094 (2020).
Allen, I. C. et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30, 556–565 (2009).
Wang, X. Q. et al. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat. Immunol. 15, 1126–1133 (2014).
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Kell, A. M. & Gale, M. Jr. RIG-I in RNA virus recognition. Virology 479, 110–121 (2015).
Zhang, H. L. et al. West Nile virus NS1 antagonizes interferon beta production by targeting RIG-I and MDA5. J. Virol. 91, e02396 (2017).
Seth, R. B., Sun, L. J., Ea, C. K. & Chen, Z. J. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappa B and IRF3. Cell 122, 669–682 (2005).
Tanji, H. et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 22, 109–115 (2015).
Zhang, Y. L., Guo, Y. J., Li, B. & Sun, S. H. Hepatitis C virus single-stranded RNA induces innate immunity via Toll-like receptor 7. J. Hepatol. 51, 29–38 (2009).
Diebold, S. S. Recognition of viral single-stranded RNA by Toll-like receptors. Adv. Drug Del. Rev. 60, 813–823 (2008).
Darnell, J. E., Kerr, I. M. & Stark, G. R. JAK-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421 (1994).
Leonard, W. J. & O’Shea, J. J. JAKS and STATS: biological implications. Annu. Rev. Immunol. 16, 293–322 (1998).
Humphray, S. J. et al. DNA sequence and analysis of human chromosome 9. Nature 429, 369–374 (2004).
Naylor, S. L. et al. Human immune interferon gene is located on chromosome 12. J. Exp. Med. 157, 1020–1027 (1983).
Kotenko, S. V. et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4, 69–77 (2003).
McGonagle, D., Sharif, K., O’Regan, A. & Bridgewood, C. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun. Rev. 19, 102537 (2020).
Xu, X. et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl Acad. Sci. USA 117, 10970–10975 (2020).
Isaacs, A. & Lindermann, J. Virus interference.1. The interferon. Proc. Natl Acad. Sci. USA 147, 258–267 (1957).
Platanias, L. C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Medrano, R. F. V., Hunger, A., Mendonça, S. A., Barbuto, J. A. M. & Strauss, B. E. Immunomodulatory and antitumor effects of type I interferons and their application in cancer therapy. Oncotarget 8, 71249–71284 (2017).
Chen, J. & Ly, H. Immunosuppression by viral N proteins. Oncotarget 8, 50331–50332 (2017).
Guo, W. et al. Diabetes is a risk factor for the progression and prognosis of COVID19. Diabetes. Metab. Res. Rev. e3319, 1–9 (2020).
Valizadeh, R., Baradaran, A., Mirzazadeh, A. & Bhaskar, L. V. K. S. Coronavirus nephropathy; renal involvement in COVID-19. J. Ren. Inj. Prev. 9, e18 (2020).
Staeheli, P., Grob, R., Meier, E., Sutcliffe, J. G. & Haller, O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8, 4518–4523 (1988).
Lindenmann, J. Resistance of mouse to mice adapted influenza A virus. Virology 16, 203–204 (1962).
Von Itzstein, M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 6, 967–974 (2007).
Huang, H. et al. COVID-19: a call for physical scientists and engineers. ACS Nano 14, 3747–3754 (2020).
Chan, W. C. W. Nano research for COVID-19. ACS Nano 14, 3719–3720 (2020).
Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034 (2020).
Bhat, T. A. et al. An animal model of inhaled vitamin E acetate and Evali-like lung injury. N. Engl. J. Med. 382, 1175–1177 (2020).
Zheng, Y. Y., Ma, Y. T., Zhang, J. Y. & Xie, X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 17, 259–260 (2020).
Li, G. & De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019NCoV). Nat. Rev. Drug Discov. 19, 149–150 (2020).
Stebbing, J. et al. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 20, 400–402 (2020).
Lipsitch, M., Swerdlow, D. L. & Finelli, L. Defining the epidemiology of Covid-19—studies needed. N. Engl. J. Med. 382, 1194–1196 (2020).
Zhang, C., Shi, L. & Wang, F. S. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol. Hepatol. 5, 428–430 (2020).
Adhikari, P. et al. Intra- and intermolecular atomic-scale interactions in the receptor binding domain of SARS-CoV-2 spike protein: implication for ACE2 receptor binding. Phys. Chem. Chem. Phys. 22, 18272 (2020).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kumar, P., Sobhanan, J., Takano, Y. et al. Molecular recognition in the infection, replication, and transmission of COVID-19-causing SARS-CoV-2: an emerging interface of infectious disease, biological chemistry, and nanoscience. NPG Asia Mater 13, 14 (2021). https://doi.org/10.1038/s41427-020-00275-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41427-020-00275-8
This article is cited by
-
Bioinspired nanotopographical design of drug delivery systems
Nature Reviews Bioengineering (2023)
-
Multimodal CTC detection using stem cell antigen-specific immunosilica particles and immunofluorescent quantum dots
NPG Asia Materials (2022)