Abstract
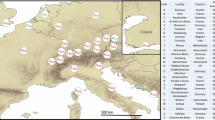
Detecting the number of invasions is crucial to understanding the process of invasion and perhaps the success of some invasive pest species. Detecting multiple invasions can be difficult using partial mitochondrial COI, however, due to lack of variation. To examine the post-invasion history of Bemisia argentifolii (also called B. tabaci Middle East-Asia Minor 1 and B biotype) in Australia and test for the presence of spatial genetic structure, we developed microsatellite loci based on the whole-genome sequence of B. argentifolii. We investigated gene flow among populations of B. argentifolii collected between 1995 and 2018, covering the time since the first detection in Australia (1994). Structure plots and PCAs of the microsatellite data revealed three clusters in 1995–1996, indicating multiple introductions. Since then, B. argentifolii has become a widespread single genetic population across the continent, with no geographic genetic structure in recent samples. The haplotype network generated from mitochondrial COI shows that Australian B. argentifolii mostly has the same haplotype as the invasive populations established elsewhere around the world. Analysis of the more recent samples showed that gene flow was high across regions, indicating movement of B. argentifolii across Australia is currently extensive. Undesirable traits and pathogens not already present in Australia, including insecticide resistance and plant viruses, could arrive with any new introductions of B. argentifolii and are likely to spread rapidly and be difficult to contain. This highlights the importance of biosecurity and continued quarantine measures to prevent new incursions, even when a species has already established.





Similar content being viewed by others
References
Bellows TS, Perring TM, Gill RJ, Headrick DH (1994) Description of a species of Bemisia (Homoptera: Aleyrodidae). Ann Entomol Soc Am 87:195–206
Bensasson D, Zhang D-X, Hartl DL, Hewitt GM (2001) Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol 16:314–321
Boykin LM, DeBarro PJ (2014) A practical guide to identifying members of the Bemisia tabaci species complex: and other morphologically identical species. Front Ecol Evol 2:1–5
Brookes DR, Hereward JP, Wilson LJ, Walter GH (2020) Multiple invasions of a generalist herbivore—Secondary contact between two divergent lineages of Nezara viridula Linnaeus in Australia. Evol Appl 13:2113–2129
Brown JK, Bird J (1992) Whitefly-transmitted geminiviruses and associated disorders in the Americas and the Caribbean Basin. Plant Dis 76:220–225
Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1010
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Chen W, Hasegawa DK, Kaur N, Kliot A, Pinheiro PV, Luan J, Stensmyr MC, Zheng Y, Liu W, Sun H, Xu Y, Luo Y, Kruse A, Yang X, Kontsedalov S, Lebedev G, Fisher TW, Nelson DR, Hunter WB, Brown JK, Jander G, Cilia M, Douglas AE, Ghanim M, Simmons AM, Wintermantel WM, Ling K-S, Fei ZJ (2016) The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol 14:110
Chu D, Li XC, Zhang YJ (2012) Microsatellite analyses reveal the sources and genetic diversity of the first-introduced Q-biotype population and the well-established B-biotype populations of Bemisia tabaci in China. Acta Entomol Sin 55:1376–1385
Chu D, Guo D, Tao Y, Jiang D, Li J, Zhang Y (2014) Evidence for rapid spatiotemporal changes in genetic structure of an alien whitefly during initial invasion. Sci Rep 4:4396
Clement M, Snell Q, Walker P, Posada D, Crandall K (2002) TCS: estimating gene genealogies. In: Proceedings 16th International Parallel and Distributed Processing Symposium, 15–19 April 2002
Costa HS, Brown JK (1991) Variation in biological characteristics and esterase patterns among populations of Bemisia tabaci, and the association of one population with silverleaf symptom induction. Entomol Exp Appl 61:211–219
Darras H, Aron S (2015) Introgression of mitochondrial DNA among lineages in a hybridogenetic ant. Biol Lett 11:20140971
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772–772
De Barro PJ (1995) Bemisia tabaci biotype B : a review of its biology, distribution and control. Technical paper (CSIRO. Division of Entomology); no. 36. CSIRO, Division of Entomology, Canberra
De Barro PJ (2011) Bemisia tabaci, the capacity to invade. In: Thompson MOW (ed) The whitefly, Bemisia tabaci (Homoptera: Aleyrodidae) interaction with geminivirus-infected host plants. Springer, Dordrecht, pp 181–204
De Barro PJ (2012) The Bemisia tabaci species complex: questions to guide future research. J Integr Agric 11:187–196
De Barro P, Ahmed M (2011) Genetic networking of the Bemisia tabaci cryptic species complex reveals pattern of biological invasions. PLoS ONE 6:e25579
De Barro PJ, Liu SS, Boykin LM, Dinsdale AB (2011) Bemisia tabaci: A statement of species status. Annu Rev Entomol 56:1–19
De Barro PJ, Scott KD, Graham GC, Lange CL, Schutze MK (2003) Isolation and characterization of microsatellite loci in Bemisia tabaci. Mol Ecol Notes 3:40–43
Delatte H, David P, Granier M, Lett JM, Goldbach R, Peterschmitt M, Reynaud B (2006) Microsatellites reveal extensive geographical, ecological and genetic contacts between invasive and indigenous whitefly biotypes in an insular environment. Genet Res 87:109–124
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B Stat Methodol 39:1–38
Dinsdale A, Cook L, Riginos C, Buckley YM, De Barro P (2010) Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann Entomol Soc Am 103:196–208
Dinsdale A, Schellhorn NA, De Barro P, Buckley YM, Riginos C (2012) Rapid genetic turnover in populations of the insect pest Bemisia tabaci Middle East: Asia Minor 1 in an agricultural landscape. Bull Entomol Res 102:539–549
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Du Z, Tang Y, He Z, She X (2015) High genetic homogeneity points to a single introduction event responsible for invasion of Cotton leaf curl Multan virus and its associated betasatellite into China. Virol J 12:163
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Food and Agriculture Organisation (2015). Australian Official Pest Reports – Absence of whitefly biotype Q in Queensland and New South Wales. Report No. AUS 23/2. International Plant Protection Convention, FAO, Rome. https://www.ippc.int/en/countries/australia/pestreports/2009/04/absence-of-whitefly-biotype-q-in-queensland-and-new-south-wales. Accessed 19 January 2020
Garnas JR, Auger-Rozenberg MA, Roques A, Bertelsmeier C, Wingfield MJ, Saccaggi DL, Roy HE, Slippers B (2016) Complex patterns of global spread in invasive insects: eco-evolutionary and management consequences. Biol Invasions 18:935–952
Gauthier N, Dalleau-Clouet C, Bouvret ME (2008) Twelve new polymorphic microsatellite loci and PCR multiplexing in the whitefly, Bemisia tabaci. Mol Ecol Resour 8:1004–1007
Gerling D, Mayer RT (1996) Bemisia: 1995: taxonomy, biology, damage, control and management. Intercept, Andover
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Gunning RV, Byrne FJ, Condé BD, Connelly MI, Hergstrom K, Devonshire AL (1995) First report of B-biotype Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Australia. J Aust ent Soc 34:116
Gunning RV (1996) B-biotype Bemisia tabaci in Australia. Paper presented at the Australian Cotton Conference, Broadbeach, Gold Coast, Queensland, August 1996
Gunning RV, Byrne FJ, Devonshire AL (1997) Electrophoretic analysis of non-B and B-biotype Bemisia tabaci (Gennaduis) (Hemiptera: Aleyrodidae) in Australia. Aust J Entomol 36:245–249
Hadjistylli M, Schwartz SA, Brown JK, Roderick GK (2014) Isolation and characterization of nine microsatellite loci from Bemisia tabaci (Hemiptera: Aleyrodidae) biotype B. J Insect Sci 14:1–3
Hedrick PW, Parker JD (1997) Evolutionary genetics and genetic variation of haplodiploids and x-linked genes. Annu Rev Ecol Evol Syst 28:55–83
Hequet E, Henneberry TJ, Nichols RL (2007) Sticky cotton: causes, effects, and prevention. Technical Bulletin 1915. U.S. Department of Agriculture-Agricultural Research Service.
Horowitz AR, Ghanim M, Roditakis E, Nauen R, Ishaaya I (2020) Insecticide resistance and its management in Bemisia tabaci species. J Pest Sci 93:893–910
Horowitz AR, Ishaaya I (2014) Dynamics of biotypes B and Q of the whitefly Bemisia tabaci and its impact on insecticide resistance. Pest Manag Sci 70:1568–1572
Hu J, De Barro P, Zhao H, Wang J, Nardi F, Liu SS (2011) An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS ONE 6:e16061
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Hulme PE (2009) Trade, transport and trouble: managing invasive species pathways in an era of globalization. J Appl Ecol 46:10–18
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Kanakala S, Ghanim M (2019) Global genetic diversity and geographical distribution of Bemisia tabaci and its bacterial endosymbionts. PLoS ONE 14:e0213946
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Lavergne S, Molofsky J (2007) Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci USA 104:3883–3888
Legg JP, Shirima R, Tajebe LS, Guastella D, Boniface S, Jeremiah S, Nsami E, Chikoti P, Rapisarda C (2014) Biology and management of Bemisia whitefly vectors of cassava virus pandemics in Africa. Pest Manag Sci 70:1446–1453
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Lester LJ, Selander RK (1979) Population genetics of haplodiploid insects. Genetics 92:1329–1345
Liu F-H, Smith SM (2000) Estimating quantitative genetic parameters in haplodiploid organisms. Heredity 85:373–382
Liu S-S, De Barro PJ, Xu J, Luan J-B, Zang L-S, Ruan Y-M, Wan F-H (2007) Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318:1769–1772
Mabvakure B, Martin DP, Kraberger S, Cloete L, van Brunschot S, Geering ADW, Thomas JE, Bananej K, Lett J-M, Lefeuvre P, Varsani A, Harkins GW (2016) Ongoing geographical spread of Tomato yellow leaf curl virus. Virology 498:257–264
Malka O, Santos-Garcia D, Feldmesser E, Sharon E, Krause-Sakate R, Delatte H, van Brunschot S, Patel M, Visendi P, Mugerwa H, Seal S, Colvin J, Morin S (2018) Species-complex diversification and host-plant associations in Bemisia tabaci: a plant-defence, detoxification perspective revealed by RNA-Seq analyses. Mol Ecol 27:4241–4256
Martinez-Carrillo JL, Brown JK (2007) Note: first report of the Q biotype of Bemisia tabaci in Southern Sonora, Mexico. Phytoparasitica 35:282–284
Meglécz E, Costedoat C, Dubut V, Gilles A, Malausa T, Pech N, Martin J-F (2010) QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics 26:403–404
Meglécz E, Pech N, Gilles A, Dubut V, Hingamp P, Trilles A, Grenier R, Martin J-F (2014) QDD version 3.1: a user-friendly computer program for microsatellite selection and primer design revisited: experimental validation of variables determining genotyping success rate. Mol Ecol Resour 14:1302–1313
Miura O (2007) Molecular genetic approaches to elucidate the ecological and evolutionary issues associated with biological invasions. Ecol Res 22:876–883
Moulton MJ, Song H, Whiting MF (2010) Assessing the effects of primer specificity on eliminating numt coamplification in DNA barcoding: a case study from Orthoptera (Arthropoda: Insecta). Mol Ecol Resour 10:615–627
Oliveira MRV, Henneberry TJ, Anderson P (2001) History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot 20:709–723
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Perring TM, Cooper AD, Rodriquez RJ, Farrar CA, Bellows TS (1993) Identification of a whitefly species by genomic and behavioral studies. Science 259:74–77
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Ridley AW, Hereward JP, Daglish GJ, Raghu S, McCulloch GA, Walter GH (2016) Flight of Rhyzopertha dominica (Coleoptera: Bostrichidae)–a spatio-temporal analysis with pheromone trapping and population genetics. J Econ Entomol 109:2561–2571
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SG (2001) The population biology of invasive species. Annu Rev Ecol Syst 32:305–332
Sattar MN, Kvarnheden A, Saeed M, Briddon RW (2013) Cotton leaf curl disease – an emerging threat to cotton production worldwide. J Gen Virol 94:695–710
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Shi X, Tang X, Zhang X, Zhang D, Li F, Yan F, Zhang Y, Zhou X, Liu Y (2018) Transmission efficiency, preference and behavior of Bemisia tabaci MEAM1 and MED under the influence of Tomato chlorosis virus. Front Plant Sci 8:2271
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved Polymerase Chain Reaction primers. Ann Entomol Soc Am 87:651–701
Song H, Buhay JE, Whiting MF, Crandall KA (2008) Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA 105:13486–13491
Subramaniam S. (2010). Development and promotion of IPM strategies for silverleaf whitefly in vegetables. Final Report, HAL Project VG05050," Rep. No. VG05050. Horticulture Australia.
Tay WT, Elfekih S, Court LN, Gordon KHJ, Delatte H, De Barro PJ (2017) The trouble with MEAM2: implications of pseudogenes on species delimitation in the globally invasive Bemisia tabaci (Hemiptera: Aleyrodidae) cryptic species complex. Genome Biol Evol 9:2732–2738
Thierry M, Bile A, Grondin M, Reynaud B, Becker N, Delatte H (2015) Mitochondrial, nuclear, and endosymbiotic diversity of two recently introduced populations of the invasive Bemisia tabaci MED species in La Réunion. Insect Conserv Divers 8:71–80
Toon A, Daglish GJ, Ridley AW, Emery RN, Holloway JC, Walter GH (2016) Random mating between two widely divergent mitochondrial lineages of Cryptolestes ferrugineus (Coleoptera: Laemophloeidae): a test of species limits in a phosphine-resistant stored product pest. J Econ Entomol 109:2221–2228
Ueda S, Brown JK (2006) First report of the Q biotype of Bemisia tabaci in Japan by mitochondrial cytochrome oxidase I sequence analysis. Phytoparasitica 34:405–411
Uller T, Leimu R (2011) Founder events predict changes in genetic diversity during human-mediated range expansions. Glob Change Biol 17:3478–3485
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3–new capabilities and interfaces. Nucleic Acids Res 40:e115
van Brunschot SL, Persley DM, Geering ADW, Campbell PR, Thomas JE (2010) Tomato yellow leaf curl virus in Australia: distribution, detection and discovery of naturally occurring defective DNA molecules. Australas Plant Pathol 39:412–423
Vyskočilová S, Tay WT, van Brunschot S, Seal S, Colvin J (2018) An integrative approach to discovering cryptic species within the Bemisia tabaci whitefly species complex. Sci Rep 8:10886–10886
Wang R, Wang J-D, Che W-N, Luo C (2018) First report of field resistance to cyantraniliprole, a new anthranilic diamide insecticide, on Bemisia tabaci MED in China. J Integr Agric 17:158–163
Ward NL, Masters GJ (2007) Linking climate change and species invasion: an illustration using insect herbivores. Glob Chang Biol 13:1605–1615
White JA, Kelly SE, Perlman SJ, Hunter MS (2009) Cytoplasmic incompatibility in the parasitic wasp Encarsia inaron: disentangling the roles of Cardinium and Wolbachia symbionts. Heredity 102:483–489
Wongnikong W, van Brunschot SL, Hereward JP, De Barro PJ, Walter GH (2020) Testing mate recognition through reciprocal crosses of two native populations of the whitefly Bemisia tabaci (Gennadius) in Australia. Bull Entomol Res 110:328–339
Acknowledgements
This research was funded by the Cotton Research and Development Corporation, Australia. We are grateful to Paul De Barro, Andras Szito, Zara Hall, Lewis Wilson, Jamie Hopkinson, Tanya Smith and Dianna Renfree for whitefly specimens that were used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. The research funders did not participate in the design or analysis of the experiments.
Ethical approval
This article does not involve any human and/or animal participants, and insects are not included in ethical consideration in Australia.
Additional information
Communicated by Rami Horowitz.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wongnikong, W., Hereward, J.P., van Brunschot, S.L. et al. Multiple invasions of Bemisia argentifolii into Australia and its current genetic connectivity across space. J Pest Sci 94, 1331–1343 (2021). https://doi.org/10.1007/s10340-021-01343-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-021-01343-w