Abstract
Mining the gene of histidine content in barley grain helps with the breeding of functional barley varieties. The study constructed a recombinant inbred lines (RILs) containing 193 families derived from the cross between Ziguangmangluoerling (ZGMLEL) (♀) and Schooner No.3 (♂). The histidine (HIS) content in the grain of the mapping population and its parents were determined by an automatic amino acid analyzer. The HIS content of ZGMLEL was 0.53 mg/g. The grain HIS content of Schooner No. 3 was 0.21 mg/g, and the grain HIS content of population ranged from 0.23 to 0.54. Genetic linkage maps, including those of seven chromosomes of barley, were constructed by using 180 pairs of simple sequence repeat (SSR) markers, with a total genetic distance of 2671.03 cM and average marker spacing of 14.84 cM. Quantitative trait locus (QTL) IciMappingV3.3 was used to analyze QTL of HIS content in barley grains, and three QTLs were detected. Mapping results showed that the three loci were located on chromosomes 2H, 4H, and 7H, respectively. The major QTL with a contribution rate of 10.11% was located on barley chromosome 4H (HVBAMMGB84-BMAG0808). The additive effect is positive (0.025). Thus, it comes from the high-value parent ZGMLEL. Another major QTL with a contribution rate of 13.75% was located on barley chromosome 7H (GBM1303-GMS056). The minor QTL with a contribution rate of 6.01% was located on chromosome 2H (Scssr03381-Scssr07759). The additive effects of 4H and 7H QTLs were negative (− 0.02 and − 0.033). So, they came from the male parent Schooner. The results provided a reference for further fine mapping, cloning, and transformation of HIS genes in barley grains.
Similar content being viewed by others
Introduction
Barley (Hordeum vulgare L.) is one of the earliest domesticated ancient cereal crops in the word (Khahani et al. 2019). Barley is a food and economic crop, as well as a medicine and food homologous crop. It is rich in minerals, lipids, and amino acids. Its grains are also rich in protein, physiologically active substances, total flavonoids, γ-aminobutyric acid (GABA), β-glucan and other nutritional functional components (Arndt et al. 2006; Xu et al. 2019). It has special efficacy and great medicinal value for the prevention and treatment of diabetes, hypertension, cardiovascular disease, chronic uremia (such as HIS), and other chronic diseases (Chan et al. 2009; Yang et al. 2020). Functional barley is one of the popular topics in barley research.
Amino acids are among the structures that make up proteins and play an irreplaceable role in most cell functions, regulation, and metabolism (Shen et al. 2019). HIS is a common amino acid that is not essential to adults, but essential to young children. So, it is called a semi-essential amino acid. HIS mainly exists in the liver and skin stratum corneum. It has a strong vasodilating effect; it can participate in the regulation of the nervous system and stimulation of the gastric acid and pepsin secretion(Gu et al. 2018; Hu et al. 2018). It has an important effect on the treatment of anemia, gastrointestinal disease, asthma, angina pectoris, cardiac insufficiency, and uremia (Lin et al. 2008; Kang et al. 2016). HIS is now used in the pharmaceutical and food industry but also with alanine co-synthesis of carnosine, which has antioxidant and anti-aging effects. Increasing the content of HIS in food is one of the important ways of histidine supplementation in humans. Thus, the production of functional meat products has become a popular research topic. As the role of HIS in the field of medicine was gradually recognized, it became one of the most commonly used drugs in medical treatment in China; so, the demand for HIS is also increasing (Sun et al. 2010).
Mining QTLs by constructing a mapping population is the main method of barley QTL research at present. Proteins and flavonoids are functional components of barley, and their QTLs have been reported (Yang et al. 2017). Agronomic trait QTLs include flowering (Ibrahim et al. 2018), disease resistance (Read et al. 2003), grain length (Watt et al. 2018), grain yield (Long et al. 2018; Falcon et al. 2019; Hu et al. 2019), spike height, spike length, spike grain number, plant height, flag leaf length, leaf area, and leaf width (Islamovic et al. 2013; Liu 2015; You 2016).
The grain amino acid content of barley has been confirmed to be a quantitative character that is affected by both gene and environment (Xu et al. 1988). Agronomic measures, meteorological factors and regional conditions under environmental conditions will affect barley’s amino acids content Nie et al. 2010; Hu et al. 2017; Li 2017; Xu et al. 2017; Bai et al. 2018). Significant progress has been made in the composition and determination of amino acid content in barley and the changes of amino acid content under different malt processing conditions (MacLeod 1951; Pomeranz et al. 1976; Williams et al. 1984; Han et al. 2009; Luo et al. 2014). For example, Nie et al. (2010) analyzed and detected the changes in the amino acid content in barley malt during germination and drying. They concluded that HIS belongs to class III (the most important) amino acid in the process of malting. It is the most important yeast metabolism.
Genetic studies have shown that barley grain amino acids were affected by endosperm triploid and maternal plant diploid genes (Yan et al. 1997). Through further research, Xu et al. (1996) found that the inheritance of HIS content in two-rowed barley grains was controlled by seed direct gene and maternal effect, and a strong interaction existed between direct dominance and the environment. However, the research on barley amino acid QTL is weak. At present, only QTLs of gamma-aminobutyric acid (GABA) content in barley grains are distributed on the chromosomes 4H, 5H, 6H and 7H (Luo et al. 2014; Yang et al. 2017). However, the QTL of HIS in barley has not been reported yet.
In the previous study, we found a variety with high HIS content in the grains (ZGMLEL), and established RILs with this variety. In this study, we used the constructed RIL mapping population as materials to analyze the correlation between HIS content in the mapping population grains and 12 morphological characters. The QTL of HIS content in barley grains were excavated. The results lay the foundation for fine mapping and transformation of histidine content genes in barley grains.
Materials and methods
Plant materials
In the winter of 2007 in Kunming, the hybrid F1 was prepared by using ZGMLEL as the female parent, and Schooner No.3 as the male parent. The F2 population was obtained by self-crossing. P1, P2 and F8 populations were planted in the experimental fields of Kunming Academy of Agricultural Sciences (altitude 1916 m) and Songming experimental fields (altitude 1902 m) from 2008 to 2011, according to the cultivation rules of RILs. The characters of F7 and F8 were stable and were considered as RILs.
Cultivation of the materials
On 28 October 2016, 193 F9 RILs lines of ZGMLEL × Schooner No.3 were planted in Yanhe Town, Yuxi City (altitude 1646 m). The previous stubble of the experimental site was rice, the soil was clay loam, and soil fertility was medium. The previous study of this project showed that the yield of regenerated wheat grain and straw was the highest at the stage of 3-leaf stage and 1 heart. In this experiment, the seedlings were cut 60 days after sowing and repeated for 2 days. The sowing rate of each plant was the same as that of the base fertilizer, that is, the sowing rate was 100 grains/m, the compound fertilizer (N: P2O5 : K2O = 13 : 5 : 7) was the base fertilizer, and the fertilizer application rate was pure N 120 kg/hm2, which emerged on November 6. Topdressing with pure N 30 kg/hm2 was performed on 2 January 2017. the other management was the same as the local conventional high-yield wheat field.
Methods
Determination of HIS content
The grain HIS content of RILs mapping population and parents was determined by Hitachi-L8900 automatic amino acid analyzer.
Investigation and determination of morphological traits
After the population plant matured, five plants were randomly selected for each repeat to measure 12 morphological traits, including available spike, invalid spike, main spike length, plant height, total length of internode and others. The mean value of each trait was calculated.
Total DNA extraction from barley
-
1.
The total DNA of leaf genome was extracted by referring to the method of Sun et al. (2009).
-
2.
DNA quality detection.
Taking the OD260/OD280 ratio between 1.8 and 2.0 as the evaluation index, the purity of DNA was detected by ultraviolet spectrophotometer (DU-7400), and the DNA concentration was calculated by using the OD260 value.
Screening of SSR markers
The 500 pairs of SSR primers used in this experiment were uniformly selected from the barley genome. The primer sequences were from http://www.wheat.pw.usda.gov/cgi-bin/graingenes, and the primers were synthesized by Beijing Dingguo Changsheng Biotechnology Co., Ltd. The synthetic primers were used to screen for polymorphism between ZGMLEL and Schooner No. 3. The selected polymorphic primers were amplified by PCR in the population.
PCR amplification and detection of reaction product
The PCR reaction system was 10 µl; 0.5 µl for upstream primer and downstream primer, 2 µl for DNA, 1.5 µl for PCR Buffer, 0.4 µl for dNTPs, 4.9 µl for ddH2O, and 0.2 µl for Taq polymerase.
PCR amplification procedure: 94 °C for 5 min, 94 °C for 40 s, 55 °C annealing for 40 s, 72 °C extension 1 min for 35 cycles, and a final extension at 72 °C for 5 min.
Detection of PCR amplification products: the amplification products were separated by denaturing 8% polyacrylamide gel electrophoresis and then detected by silver staining. The specific reagents are mentioned in the study of Sun et al. (2009).
SSR marking data record
We placed the silver-stained gel under the film observation lamp to read the tape with reference to Yang et al. (2017). We recorded it with QTL IciMapping software. If the band type was the same as the female ZGMLEL recorded “0,“ and if it is same as the father’s choice recorded “2,“ the hybrid band is recorded as “1” and the missing band is recorded as “− 1.“
QTL mapping of amino acids in grains of RILs
We used QTL IciMappingV3.3 to construct the molecular genetic linkage map of barley RIL population and used the Inclusive Composite Interval Mapping (ICIM) method to map QTL. Using LOD ≥ 2.5 as the threshold of QTL existence, We analyzed the linkage relationship between histidine content and marker genotypes in the mapping population, determined the relative position of histidine QTLs on chromosomes, and estimated their genetic effects (Wang 2009). The nomenclature of QTL followed the method of McIntosh (2013).
Results and analysis
Analysis of the variation of the grain HIS content in RILs
The grain HIS frequency map and variation table of ZGMLEL RILs are shown in Fig. 1; Table 1. Combined with Fig. 1; Table 1, the population kurtosis was − 0.73, and the population deviation value was − 0.59, indicating that the HIS content distribution in the population grain was the slope peak (left-deviated normal distribution). The strains with lower HIS content accounted for a larger proportion in the population, and the intermediate HIS content lines were relatively less in the population. According to Table 1, the average HIS content of the population was between the two parents. The HIS content of the male parent Schooner No.3 was 0.21 mg/g, which was a low-value parent, and the HIS content of the female parent ZGMLEL was 0.53 mg/g, which was a high-value parent. The HIS content in population grains ranged from 0.23 mg/g to 0.54 mg/g, with the average value of was 0.38 ± 0.01 mg/g, and the coefficient of variation was 1.90%. The average value of the population was higher than that of the parents, and the content of HIS was higher than that of the strains with high parent content, indicating that the HIS content showed a trend of high parental segregation in the RIL mapping population.
Correlation analysis between grain HIS content and 12 morphological traits in RILs
The correlation between HIS content in population grain and 12 morphological characters is presented in Table 2. The HIS content was significantly or very significantly correlated with six traits. Significant negative correlation was found with available spike and panicle weight per plant. Extremely significant positive correlation was found with invalid spike and blighted grains. Extremely significant negative correlation was found with number of solid grains and setting rate. Further analysis showed that the HIS content in the grain was most closely related to setting rate, and the correlation coefficient was − 0.34. The correlation between histidine and 12 morphological traits showed that the morphological traits of histidine and barley yield were significant or negative. Thus, increasing barley grain yield might reduce histidine content.
SSR marker analysis of ZGMLEL RILs population
-
(1)
SSR marker analysis
Among the 500 pairs of SSR primers, 180 could obtain clear and polymorphic bands, and the segregation of the above markers in the population was basically consistent with the Mendel segregation ratio of 1:1, which was a homozygous locus.
-
(2)
Construction of genetic map
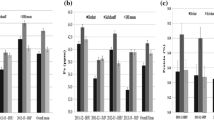
We used 180 pairs of primers with good polymorphism between parents to conduct population amplification and constructed seven linkage groups. These seven linkage groups were constructed (Fig. 2). The related information of the linkage groups was sorted out in Table 3. According to the Table 3, the total length of the genetic linkage map was 2671.03 cM, and the average genetic distance was 14.84 cM. The longest covering distance was found in the 7H chromosome, which was 542.89 cM, and the number of markers was the highest (39). The average genetic distance between markers was 13.92 cM. The shortest distance was 233.72 cM on chromosome 6H, and number of markers was the lowest (15). The average distance between markers was 15.59 cM.
QTL analysis of grain HIS content in the ZGMLEL RILs
The QTL information of grain HIS content in ZGMLEL RILs is shown in Fig. 3 and Table 4. According to the Table 4, three QTLs related to the HIS content in barley grains were excavated; these were located on 2H, 4H and 7H chromosomes, which could explainthe genetic variations of 6.01%, 10.11% and 13.75%, respectively. We initially named them QHIS. KAAS-2H, QHIS. KAAS-4H and QHIS. KAAS-7H. Among the three QTLs, QHIS. KAAS-4H and QHIS. KAAS-7H were regarded as major QTLs. They were located in the marker HVBAMMGB84-BMAG0808 and GBM1303-GMS05. QHIS. KAAS-4H had additive effect from female parent ZGMLEL 0.025, QHIS. KAAS-7H had additive effect from male Schooner No.3 0.033; QHIS. KAAS-2H was regarded as a minor QTL located between marker Scssr03381 and Scssr07759 and had additive effect from Schooner No.3 0.02. In summary, the grain HIS content of barley is a quantitative character controlled by both major and minor genes.
Discussions
Barley is listed as a health food by the United States Department of Agriculture (Newman and Newman 2008). Whole-grain food made from barley is considered to be “healthy food” for humans and plays an important role in preventing chronic diseases such as cardiovascular disease and cancer (Gangopadhyay et al. 2015; Qi 2018). The study on the correlation between grain HIS content and 12 morphological characters in barley showed the significant negative correlation between grain HIS content and barley yield. Thus, increasing barley yield may decrease grain HIS content.
Using the RIL population to make a genetic mapping and mining QTL can analyze the additive effect of QTLs, This population is suitable for plant QTL mapping. Many barley QTLs were obtained by using this population. For example, Lu (2015) used the RIL population constructed by barley (GP × H602) as a material for genetic linkage map and QTL analysis of seven agronomic traits, and a total of 28 QTLs were detected. Hu et al. (2017) used the RIL mapping population constructed from barley cultivar Baudin and germplasm material CN4079 to map the traits related to phosphorus efficiency at tillering stage in barley under different phosphorus levels, and 16 QTLs loci were detected. Tamang et al. (2019) discovered 12 QTLs controlling the Spot Form Net Blotch in barley by using the Tradition × Pinnacle RILs mapping population. Yang et al. (2019) detected two QTLs related to resistant starch in 2 years by using the RILs mapping population constructed by “Yunpi No. 2” and “Dali Barley” as materials.
In this study, we used ZGMLEL × Schooner No.3 RIL mapping population to map three QTLs of HIS content in barley grains, which were located on chromosomes 2H, 4H, and 7H, respectively. In previous studies, QTLs of other traits were found in barley 2H, 4H, and 7H Emebiri et al. 2005; Li 2007; Nie et al. 2010; Guo 2012; Yu 2014; Liu 2015; Wang 2016; Yang et al. 2017; Fei 2018). However, no reports on histidine QTL in barley grains are available. So, the three QTLs found in this study were all new QTLs controlling histidine content in barley grains. Further analysis revealed that the locus found in this study on the 4H chromosome had common markers with the protein sites reported by Yang et al. (2017) on 4H (BMAG0808). This result may be due to the fact that the mapping populations used in this study are consistent with those used by Yang. Amino acids are the basic units of proteins. Therefore, a certain correlation may exist between QTLs controlling protein content and histidine content in barley grains.. We speculated that there might be a certain correlation might exist between the QTL controlling the protein content in barley grains and the QTL controlling the histidine content in barley grains.
References
Arndt EA (2006) Whole grain barley for today’s health and wellness needs. Cereal Foods World 51(1):20–22. https://doi.org/10.1094/CFW-51-0020
Bai YX, Yao XH, Yao YH, Yang LN, Wu KL (2018) Effects of water deficit on the contents of N, P and K and amino acids accumulation in wheat grains of Qinghai-Tibet Plateau. J China Agric Univ 23(07):11–18. https://doi.org/10.11841/j.issn.1007-4333.2018.07.02
Chan CF, Chan WK, Sze MY (2009) The effects of β-glucan on human immune and cancer cells. J Hematol Oncol 2(6):25–36. https://doi.org/10.1186/1756-8722-2-25
Emebiri LC, Moody DB, Horsley R, Panozzo J, Read BJ (2005) The genetic control of grain protein content variation in a doubled haploid population derived from a cross between Australian and North American two-rowed barley lines. J Cereal Sci 41(1):107–114. https://doi.org/10.1016/j.jcs.2004.08.012
Falcon CM, Horsley R, Hu G, Blake T, Smith KP (2019) Mapping QTLs for grain protein concentration and agronomic traits under different nitrogen levels in barley. Crop Sci 59(1):68–83. https://doi.org/10.2135/cropsci2018.03.0208
Fei XR, Zhu J, Guo H, Lv C, Guo BJ, Xu RG (2018) Genetic analysis and QTL mapping of resistance to yellow mosaic disease in barley (Hordeum vulgare L.). J Triticeae Crops 038(001): 1–7; https://doi.org/10.7606/j.issn.1009-1041.2018.01.01
Gu P, Zhang G, Deng Z et al (2018) A novel label-free colorimetric detection of L-histidine using Cu2+-modulated G-quadruplex-based DNA zymes. Spectrochim Acta Part A 203:195–200. https://doi.org/10.1016/j.saa.2018.05.084
Guo LL (2012) Construction of molecular genetic map and QTL mapping for main agronomic charaters and functional compounds of barley. Sichuan Agricultural University, Sichuan, pp 32–33
Gangopadhyay N, Hossain MB, Rai DK, Brunton NP (2015) A review of extraction and analysis of bioactives in oat and barley and scope for use of novel food processing technologies. Molecules 20(6):10884–10909. https://doi.org/10.3390/molecules200610884
Han L, Yun JM, Han XF (2009) Determination of amino acids, major minerals and vitamin in barley malt (Hordeum ‘Ganpi No. 3’). J Gansu Agric Univ 44(3):151–152
Hu DY, Cai L, Chen GD, Zhang XZ, Liu CJ (2017) Mapping QTLs for phosphorus efficiency at tillering stage under different phosphorus levels in barley (Hordeum vulgare L.). Acta Agron Sin 43(12):1746–1759
Hu M, Wu SG, Wang J, Qi GH, Zhang HJ (2018) Physiological function and application of histidine in animal production. China Feed 07:38–45. https://doi.org/10.15906/j.cnki.cn11-2975/s.20180708
Hu H, Ahmed I, Choudhury S et al (2019) Wild barley shows a wider diversity in genes regulating heading date compared with cultivated barley. Euphytica 215:75. https://doi.org/10.1007/s10681-019-2398-1
Ibrahim A, Harrison M, Meinke H, Fan Y, Johnson P, Zhou MX (2018) A regulator of early flowering in barley (Hordeum vulgare L.). PLoS ONE 13(7):e0200722. https://doi.org/10.1371/journal.pone.0200722
Islamovic E, Obert DE, Oliver RE et al (2013) A new genetic linkage map of barley (Hordeum vulgare L.) facilitates genetic dissection of height and spike length and angle. Field Crops Res 154:91–99. https://doi.org/10.1016/j.fcr.2013.06.001
Kang P, Zhang Y, Liu S, Wang ZW, Chen T (2016) Research on L-histidine production by escherichia coli. Food Ferment Ind 42(11):249–254. https://doi.org/10.13995/j.cnki.11-1802/ts.201611043
Khahani B, Tavakol E, Shariati VJ (2019) Genome-wide meta-analysis on yield and yield-related QTLs in barley (Hordeum vulgare L.). Mol Breed, 39 (4): 56; https://doi.org/10.1007/s11032-019-0962-y
Li Z (2007) QTL analysis of plant height and yield component traits in barley. Chinese Academy of Agricultural Sciences, Beijing, pp 26–30
Li N, Ruan JJ, Yan J, Zhao G, Dai YL, Chen JP (2017) Effect of genotype and environment on nutritional traits of wild barley (Hordeum spontaneum) from Israel. J Triticeae Crops 37(12):1564–1570. https://doi.org/10.7606/j.issn.1009-1041.2017.12.06
Lin YY (2008) Establishment of a method for the determination of histidine. Occup Health 8:734–736. https://doi.org/10.3969/j.issn.1004-1257.2008.08.012
Liu LP (2011) QTL mapping for important traits of flag leaf in barley. Huazhong Agricultual University, Huazhong, pp 27–34
Liu ZH, Holmes DJ, Faris JD, Chao S, Brueggeman RS, Edwards MC (2015) Necrotrophic effector-triggered susceptibility (NETS) underlies the barley–Pyrenophora teres f. teres interaction specific to chromosome 6H. Mol. Plant Pathol 16(2):188–200. https://doi.org/10.1111/mpp.12172
Long Z, Jia Y, Tan C et al (2018) Genetic mapping and evolutionary analyses of the black grain trait in barley. Front Plant Sci 9:1921. https://doi.org/10.3389/fpls.2018.01921
Lu WY (2015) Construction of genetic linkage map and detection of QTLs for the agronmic traits in barley and gene mapping of Lesion resembing disease mutant bspl1. Hangzhou Normal University, Hangzhou, pp 24–32
Luo XJ, Guo LL, Yang XY, Liu XC, Yu CL, Qi GC (2014) QTL mapping of γ-Aminobutyric acid content in grains of barley. Southwest China J Agric Sci 27(03):950–954. https://doi.org/10.3969/j.issn.1001-4829.2014.03.009
MacLeod AM (1951) Sugars and amino acids of the barley grain. J Inst Brew 57(3):163–169. https://doi.org/10.1002/j.2050-0416.1951.tb01619.x
McIntosh RA, Yamazaki Y, Dubcovsky J, Rogers J, Morris C, Appels R, Xia XC (2013) Catalogue of gene symbols for wheat. CD, In: Proceedings of the 12th international wheat genetics symposium, Yokohama, Japan, pp 13–14
Newman RK, Newman CW (2008) Barley for food and health: Science, technology, and products, pp 186–187. https://www.ovid.com/product-details.9319.html
Nie C, Wang CL, Zhou GT, Dou FJ, Huang MM (2010) Effects of malting conditions on the amino acid compositions of final malt. Afr J Biotechnol, 9(53): 9018–9025; https://doi.org/10.3969/j.issn.0254-5071.2010.10.027
Pomeranz Y, Robblns GS, Gilbertson JT (1976) Protein content and amino acid composition of tissue components in developing barley. J Food Sci 41(2):283–285. https://doi.org/10.1111/j.1365-2621.1976.tb00601.x
Qi WT, Chen WR, Chen YJ, Yun TT, Sang W, Ren GX (2018) An exploration of the correlation between the contents of functional active substances in barley and their antioxidant capacity. J Chin Inst Food Sci Technol 18(11):238–245
Read BJ, Raman H, Mcmichael G, Chalmers KJ, Ablett GA, Platz GJ, Raman R, Genger RK, Boyd WJR, Li CD, Grime CR, Park RF, Wallwork H, Prangnell R, Lance RCM (2003) Mapping and QTL analysis of the barley population Sloop × Halcyon. Aust J Agric Res 54(11):1155–1162. https://doi.org/10.1071/AR03037
Shen R, Zou L, Wu S, Li T, Wang J, Liu J, Ling L (2019) A novel label-free fluorescent detection of histidine based upon Cu2+-specific DNAzyme and hybridization chain reaction. Spectrochim Acta Part A 213:42–47. https://doi.org/10.1016/j.saa.2019.01.062
Sun ZH, Zeng YW, Yang SM, Pu XY, Du J (2009) Identification of QTLs about zinc content in brown rice for near-isogenic lines for Towada. Mol Plant Breed 7(02):264–268. https://doi.org/10.3969/j.issn.1672-416X.2009.02.007
Sun XY, Yang PP, Li X, Wang Y (2010) Study Progress of fermentation conditions of L-histidine and screening of high productive strain. China Brew 9:28–30. https://doi.org/10.3969/j.issn.0254-5071.2010.09.009
Tamang P, Richards JK, Alhashal A, Poudel RS, Horsley RD, Friesen TL, Brueggeman RS (2019) Mapping of barley susceptibility/resistance QTL against spot form net blotch caused by Pyrenophora teres f. maculata using RIL populations. Theor Appl Genet 132(7):1953–1963. https://doi.org/10.1007/s00122-019-03328-x
Wang JK (2009) Inclusive composite interval mapping of quantitative trait genes. Acta Agron Sin 35(02):239–245. https://doi.org/10.3724/SP.J.1006.2009.00239
Wang JB (2016) Genetic analysis of some grain yield and quality related traits in barley. Huazhong Agricultural University, Huazhong, pp 29–34
Watt C, Zhou G, Mcfawn LA, Chalmers KJ, Li C (2018) Fine mapping of qGL5H, a major grain length locus in barley (Hordeum vulgare L.). Theor Appl Genet, 132 (4): 883–893; https://doi.org/10.1007/s00122-018-3243-y
Williams PC, Preston KR, Norris KH, Starkey PM (1984) Determination of amino acids in wheat and barley by near-infrared reflectance spectroscopy. J Food Sci 49(1):17–20. https://doi.org/10.1111/j.1365-2621.1984.tb13658.x
Xu SY, Yan XF, Xu ZR, Zhu J (1996) Genetic analysis of nonessential amino acid contents in two-rowed barley. J Zhejiang Agric Univ 6:19–25. https://doi.org/10.3321/j.issn:1008-9209.1996.06.006
Xu SY, Chen HJ, Guo YY, Ding SR (1988) Relationship between amino acid content in barley seeds and cultivars and cultivation conditions. Zhejiang Agric Sci 6:47–47. https://doi.org/10.3969/j.issn.0254-5071.2010.09.009
Xu SJ, Zhang FY, Liu ZP et al (2017) Effects of sowing DATE and nitrogen application on grain protein content and free amino acid content during grain filling in spring barley. J Triticeae Crops 37(12):1611–1618. https://doi.org/10.7606/j.issn.1009-1041.2017.12.12
Xu WL, Dong WZ, Wang J, Xu J, Ma Y, Sha YS (2019) Research progresses on the nutrients of barley grain and their utilization. Barley Cereal Sci 16:40–42
Yan XF, Li WF (1997) Analysis of seed and maternal genetic effects on the contents of seven essential amino acids in two -rowed barley. Chin Agric Sci 30(2):34–41
Yang XM, Du J, Zeng YW, Pu XY, Yang SM, Yang T, Wang LX, Yang JZ (2017) QTL mapping of protein and related functional components content in barley grains. Sci Agric Sin 50(2):205–215. https://doi.org/10.3864/j.issn.0578-1752.2017.02.001
Yang T, Yang JZ, Luo HM, Du J, Pu XY, Yang XM, Li X, Zeng YW (2019) QTL analysis of resistant starch content in barley grain using the population of recombinant inbred lines. Mol Plant Breed 17(8):1032
Yang XM, Li X, Pu XY, Du J, Ali MK, Yang JZ, Zeng YW, Yang T (2020) QTL mapping for total grain anthocyanin content and 1000-kernel weight in barley recombinant inbred lines population. Acta Agron Sin 46(1):52–61. https://doi.org/10.3724/SP.J.1006.2020.91024
You J (2016) QTL mapping of kernal and plant type using a RILs population in barley. Sichusn Agricultual University, Sichusn, pp 20–22
Yu CL (2014) QTL mapping for four agronomic and main flavonoid contents. Sichuan Agricultural University, Sichusn, pp 32–36
Funding
The research and publication of this article was funded by China Agriculture Research System (CARS-05-01A-04), the National Natural Science Foundation of China (31260326), International Cooperation Project of Yunnan (2019IB011), Key R & D Plan of Yunnan (2018BB013), Science and Technology Benefiting People Program of Yunnan (2016RA032), and R & D Funding Input Subsidy Project of Yunnan and Three-year Plan for Yunnan (2018DS004).
Author information
Authors and Affiliations
Contributions
Data analysis: [ZS], [LY], and [XY]; Investigation: [JD], [XP]; Methodology: [ZS], [YZ], [XY], [JD], [XP], and [JY]; Project administration: [ZS], and [YZ]; Software: [YZ], [XY], and [JY]; Writing – original draft: [LY]; Writing – review and editing: [ZS] and [YZ].
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaomeng Yang have contributed equally to this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yan, L., Yang, X., Sun, Z. et al. Analysis and mapping quantitative trait loci for histidine content in barley (Hordeum vulgare L.) using microsatellite markers. Genet Resour Crop Evol 68, 2107–2118 (2021). https://doi.org/10.1007/s10722-021-01121-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01121-8