Abstract
The heat-stable peptidase AprX, secreted by psychrotolerant Pseudomonas species in raw milk, is a major cause of destabilization and premature spoilage of ultra-high temperature (UHT) milk and milk products. To enable rapid detection and quantification of seven frequent and proteolytic Pseudomonas species (P. proteolytica, P. gessardii, P. lactis, P. fluorescens, P. protegens, P. lundensis, and P. fragi) in raw milk, we developed two triplex qPCR assays taking into account species-dependent differences in AprX activity. Besides five species-specific hydrolysis probes, targeting the aprX gene, a universal rpoB probe was included in the assay to determine the total Pseudomonas counts. For all six probes, linear regression lines between Cq value and target DNA concentration were obtained in singleplex as well as in multiplex approaches, yielding R2 values of > 0.975 and amplification efficiencies of 85–97%. Moreover, high specificity was determined using genomic DNA of 75 Pseudomonas strains, assigned to 57 species, and 40 other bacterial species as templates in the qPCR. Quantification of the target species and total Pseudomonas counts resulted in linear detection ranges of approx. 103–107 cfu/ml, which correspond well to common Pseudomonas counts in raw milk. Application of the assay using 60 raw milk samples from different dairies showed good agreement of total Pseudomonas counts calculated by qPCR with cell counts derived from cultivation. Furthermore, a remarkably high variability regarding the species composition was observed for each milk sample, whereby P. lundensis and P. proteolytica/P. gessardii were the predominant species detected.
Key points
• Multiplex qPCR for quantification of seven proteolytic Pseudomonas species and total Pseudomonas counts in raw milk
• High specificity and sensitivity via hydrolysis probes against aprX and rpoB
• Rapid method to determine Pseudomonas contamination in raw milk and predict spoilage potential
Similar content being viewed by others
Introduction
Premature spoilage of ultra-high temperature (UHT) milk and milk products due to microbial extracellular enzymes is challenging for the dairy industry, both from an economic and a technical point of view (Hantsis-Zacharov and Halpern 2007; Marchand et al. 2009b; Stoeckel et al. 2016a, b; von Neubeck et al. 2016). Cold storage of raw milk before processing favors the growth of psychrotolerant bacteria, especially Pseudomonas, which soon dominate the microbiota (Lafarge et al. 2004; De Jonghe et al. 2011; von Neubeck et al. 2015). Several Pseudomonas species produce the extracellular, caseinolytic peptidase AprX, which is heat stable and remains partly active even after UHT treatment. Residual AprX activity can then cause negative effects in milk, such as off-flavors, particle formation, fat separation, or age gelation, all leading to instability and shelf life reduction of processed dairy products (McKellar 1981; Sørhaug and Stepaniak 1997; Matéos et al. 2015; Stoeckel et al. 2016a; Marchand et al. 2017).
The alkaline zinc-metallopeptidase AprX, belonging to the serralysin protease family, has a molecular weight of 45–50 kDa and is encoded by the polycistronic aprX-lipA2 operon (Schokker and van Boekel 1997; Woods et al. 2001; Marchand et al. 2009b). For many aprX-possessing Pseudomonas species, this operon additionally includes genes coding for a peptidase inhibitor (AprI), an ABC-transport system (AprDEF), two putative autotransporters (PrtA and PrtB), and a lipase (LipA) (Duong et al. 2001; Woods et al. 2001; Maier et al. 2020). Several studies revealed a high variability of milk-associated Pseudomonas species and strains regarding their proteolytic potential, which has been proposed to be due to different gene expression and regulation mechanisms (Dufour et al. 2008; Marchand et al. 2009b; Bagliniere et al. 2012; von Neubeck et al. 2015; Caldera et al. 2016). However, genetic variations also seem to play a role, as aprX gene sequences of Pseudomonas spp. isolated from raw milk were shown to be very heterogeneous (Marchand et al. 2009b). Moreover, different aprX-lipA2 operon structures were identified in the genus and a correlation between the type of operon organization and the proteolytic potential of pseudomonads was observed (Maier et al. 2020). Regarding the occurrence in raw milk, Pseudomonas proteolytica, Pseudomonas lundensis, Pseudomonas lactis, Pseudomonas fragi, Pseudomonas protegens, Pseudomonas gessardii, and Pseudomonas fluorescens were found to be the most frequent species, revealing various proteolytic capacities. While strains of P. proteolytica, P. lactis, P. protegens, P. gessardii, and P. fluorescens exhibited mainly high proteolytic activity, isolates of P. lundensis or P. fragi had middle or low proteolytic potential (Marchand et al. 2009a; De Jonghe et al. 2011; Baur et al. 2015; von Neubeck et al. 2015; Caldera et al. 2016; Glück et al. 2016; Maier et al. 2020).
Sensitive and rapid applications for determination of milk-spoiling Pseudomonas strains or AprX amounts are required to control raw milk quality and avoid deterioration of processed dairy products. Time-consuming culturing on selective media is not suitable to predict the spoilage potential of raw milk samples prior to processing. Regarding molecular methods, only a few immunological assays with monoclonal antibodies directed against single AprX proteins of specific Pseudomonas strains have been developed, which are not appropriate for a broader application in raw milk containing multiple species (Birkeland et al. 1985; Clements et al. 1990; Matta et al. 1997). Moreover, PCR-based approaches have been performed using aprX as a target gene to indirectly detect the spoilage potential (Martins et al. 2005; Marchand et al. 2009b; Machado et al. 2013). However, these methods were applied in pasteurized, reconstituted, or sterilized milk and are not sensitive enough to be used in raw milk, having a lower detection limit of, e.g., 107 colony-forming units (cfu) per ml (Machado et al. 2015). Also, most former molecular assays focused on the aprX or peptidase detection of P. fluorescens strains, neglecting other common milk-spoiling species, such as members of the P. gessardii and P. fragi subgroup.
Consequently, until now, there is no genetic method to discriminate between distinct Pseudomonas species with various proteolytic activity present in raw milk. Thus, the aim of this study was to develop a species-specific multiplex qPCR assay, able to quantify seven of the most frequent and proteolytic Pseudomonas species in raw milk as well as the total Pseudomonas counts. Overall, two triplex assays were established using species-specific probes, targeting aprX gene sequences, and one universal rpoB probe, directed against all members of the genus.
Material and methods
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in the Supplementary Table S1. In total, 75 strains of 57 Pseudomonas species and isolates of 40 other bacterial species, belonging to 25 different genera, were chosen. Among them, 61 strains originated from raw milk, 18 from environmental samples, 9 from milk or semi-finished milk products, 8 from water, 8 from soil, 4 from food environments, and 4 from human samples. For cultivation, most bacterial strains were grown under aerobic conditions on TS-agar (Carl Roth GmbH, Karlsruhe, Germany) at 30 °C for 24–96 h. Bifidobacterium longum was cultivated under anaerobic conditions at 37 °C on TOS-agar (Merck KGaA, Darmstadt, Germany). Members of Lactobacillus, Leuconostoc, and Lactococcus were grown under anaerobic conditions at 30 °C on APT-agar for 48 h (Merck KGaA, Darmstadt, Germany). Overnight cultures of Pseudomonas spp. were performed by inoculating 4 ml tryptic soy broth (TSB, Merck KGaA, Darmstadt, Germany) with cell material from one colony and incubated at 30 °C and 150 rpm for 16 h. Cell counts were determined on TS-agar (total cell count) as well as on selective CFC-agar (Pseudomonas cell count, Merck KGaA, Darmstadt, Germany) after incubation at 30 °C for 24 h.
Raw milk samples
For spiking experiments, fresh raw milk was obtained from a test farm of TUM (Forschungsstation Veitshof, Freising, Germany) and stored at − 20 °C until use, to ensure constant experimental conditions. For validation of the qPCR assay, 60 raw milk samples from 13 different dairies located all over Germany were analyzed. All samples were shipped refrigerated for 1–3 days. Total and Pseudomonas cell counts of raw milk samples were determined immediately after receipt, and the remaining samples were stored at − 20 °C until further processing.
Bacterial DNA extraction from raw milk samples
Bacterial DNA was extracted from raw milk samples using the DNeasy® PowerFood® Microbial Kit (Qiagen N.V., Hilden, Germany) combined with an EDTA pre-treatment. In brief, raw milk samples of 7.2 ml (4 × 1.8 ml) were centrifuged (2 min, 16000g, room temperature (RT)), and the supernatants and the covering fat layers were removed. The remaining pellets were then resuspended and united in a total of 1 ml ¼ Ringer’s solution (Merck KGaA, Darmstadt, Germany). After adding 300 μl EDTA (0.5 M) and 200 μl 1× TE-buffer, samples were incubated (1 min, RT), centrifuged (2 min, 16000g, RT), and the supernatants were carefully removed. Bacterial DNA in the remaining pellets was subsequently isolated following the manufacturer’s instructions of the kit. DNA was eluted in 50 μl elution buffer and stored at − 20 °C until use.
Reconstruction of aprX and rpoB single-gene phylogenies
Protein-coding genes of 61 Pseudomonas strains were predicted based on NCBI genome assemblies (Supplementary Table S2) using Prodigal v2.6 (Hyatt et al. 2010). AprX and rpoB gene sequences were extracted from gene predictions by searching for unidirectional best BLASTp v2.2.25+ (Camacho et al. 2009) hits to the NCBI reference sequences with GenBank identifiers AGL85010.1 and KKJ93525.1, respectively. Subsequently, multiple sequence alignments were calculated with ClustalW (Thompson et al. 1994) and used for maximum likelihood phylogenetic tree reconstruction via the MEGAX v10.0.5 software (Kumar et al. 2018) applying the general time reversible (GTR) model under the assumption of rate heterogeneity (+G) and a proportion of invariant sites (+I). To infer branch confidence values, 500 bootstrap replicates were computed for each tree. Finally, both phylogenies were visualized using the interactive Tree Of Life (iTOL) v5.3 online tool (Letunic and Bork 2019). The strain P. aeruginosa WS 5022 served as outgroup to root the trees.
Estimation of aprX and rpoB sequence similarities
Pairwise p-distances of aprX and rpoB gene sequences from Pseudomonas strains were calculated based on single-gene multiple sequence alignments using the MEGAX v10.0.5 software (Kumar et al. 2018). After subtracting distance values from 1 and multiplication by a factor of 100, pairwise sequence similarities were obtained. The outgroup strain P. aeruginosa WS 5022 was excluded from the comparison.
Primer and probe design
All primer and hydrolysis probes used in this study and their main characteristics are listed in Table 1. In total, five species-specific hydrolysis probes and primers targeting aprX were created to detect the following species: P. proteolytica, P. gessardii and P. gessardii-like species (probe 1; P1); P. fluorescens, P. lactis and P. lactis-like species (probe 2; P2); P. protegens and P. protegens-like species (probe 3; P3); P. fragi (probe 4; P4); P. lundensis and P. lundensis-like species (probe 5; P5). Additionally, one universal rpoB probe (probe 6; P6) and primer pair, targeting all members of Pseudomonas, were produced. For design, aprX and rpoB sequences of 61 Pseudomonas strains (30 type strains and 31 environmental isolates) were selected. All isolates and associated genome accession numbers are listed in the Supplementary Table S2. Respective sequences were aligned applying MEGA X (Kumar et al. 2018) and conserved regions were identified manually and checked for suitability. The formation of self- and cross-dimers of primers and probes was analyzed using Multiple Primer Analyzer (Thermo Fisher Scientific Inc., Waltham, Massachusetts), and hairpin formation was tested via OligoCalc (Kibbe 2007). Resulting primers had an annealing temperature between 55.2 and 58.2 °C, a GC content of 47.6 to 64.7%, and a length of 17 to 21 nucleotides. Hydrolysis probes revealed an annealing temperature between 63.1 and 65.8 °C, a GC content of 56.5 to 68.4%, and a length between 19 and 23 nucleotides. All oligonucleotides and hydrolysis probes were obtained from Eurofins Genomics Germany GmbH (Ebersberg, Germany).
qPCR optimization and conditions
Quantitative PCR was performed with the real-time PCR detection system CFX96/C1000 TouchTM (Bio-Rad Laboratories Inc., Hercules, CA, USA) using the CFX MaestroTM software and the following reaction conditions: Initial denaturation step at 95 °C for 2 min and 35 cycles including denaturation at 95 °C for 5 s and annealing/extension at 61 °C for 15 s. Optimal primer concentration was determined separately for each primer pair via a SYBR green qPCR assay. For this, a total reaction volume of 10 μl was used, including 5 μl SsoAdvancedTM Universal SYBR® Green Supermix (Bio-Rad Laboratories Inc., Hercules, CA, USA), 1 μl per primer in different concentrations (200 nM, 400 nM, or 600 nM), and 1 μl target DNA. The optimal quantity of the hydrolysis probes was subsequently determined by applying different probe concentrations (150 nM, 200 nM, 250 nM) with the previously defined primer concentrations in singleplex probe-based qPCR. Singleplex and multiplex probe-based qPCR was performed in 10 μl reaction volume, containing 5 μl of the SsoAdvanced™ Universal Probes Supermix (Bio-Rad Laboratories Inc., Hercules, CA, USA), primers, and probe in optimized concentrations (Table 1) and 1 or 2 μl DNA template, depending on the experiment. Multiplex qPCR utilizing six probes was split into two triplex assays, containing each three hydrolysis probes and the respective primer pairs as listed in Table 1.
Production of artificial DNA mixtures
In order to evaluate the multiplex qPCR assays, complex DNA pools were generated to be used as templates. Two strains per target species of each species-specific probe (P1–P5) were selected as representatives. Strains were grown on TS-agar plates for 24 h, and DNA was extracted using the QIAamp DNA Mini Kit (Qiagen N.V., Hilden, Germany) according to manufacturer’s specifications. Then, tenfold dilution series containing 100 to 0.01 ng/μl DNA of each strain were prepared and concentrations were checked using the Qubit 2.0 Fluorometer (Invitrogen AG, Carlsbad, CA, USA). Subsequently, identical concentrations of five target DNAs, each of which is detected by one of the five species-specific probes, were combined in a single DNA pool. In total, six distinct DNA mixtures (pool 1–6) with various target DNA compositions were prepared, which are summarized in Supplementary Table S3. In addition, each DNA mixture was generated in five different total DNA concentrations (0.01, 0.1, 1.0, 10, and 100 ng/μl) using the dilution series of the single target DNAs. Thus, the final concentration of target DNAs per species-specific probe (P1–P5) was between 0.002 and 20 ng/μl in the DNA pools. With regard to the universal rpoB probe (P6), detecting all pseudomonads, target DNA concentration ranged from 0.01 to 100 ng/μl in the DNA mixtures.
Amplification efficiency and sensitivity of the qPCR assays
For each hydrolysis probe, singleplex qPCR was performed before conducting multiplex qPCR, in order to check for probe functionality and possible interfering interactions between primer and probes. For both single- and multiplex qPCR, all six DNA mixtures (Supplementary Table S3) were used in different concentrations as templates. The quantification cycle (Cq) values obtained per probe from differing target DNA concentrations in the singleplex approach were compared with corresponding Cq values received from multiplex qPCR. For the determination of reaction efficiencies, regression lines were created by plotting the Cq values versus the log of the target DNA concentration used for qPCR. The amplification efficiency (E) was calculated for each probe from the slopes using the formula: \( E={10}^{\frac{-1}{\mathrm{slope}}}-1 \)
In order to evaluate the sensitivity of the qPCR assay, the linear dynamic range was determined as well as the lower limit, defined as the number of detectable gene copies when applying the minimum target DNA concentration (0.002 ng for probes P1–P5 and 0.01 ng for probe P6). Copy numbers of aprX (for P1–P5) and rpoB (P6) were calculated for each probe separately, using the following formula (Staroscik 2011-2020):
As aprX and rpoB present single-copy genes, the genome number is equivalent to the number of gene copies. Genome sizes of 18 target strains, which were used for the production of the DNA mixtures, were taken from NCBI and averaged per probe: 6265591 bp (P1), 6528297 bp (P2), 6799673 bp (P3), 5227135 bp (P4), 5131361 bp (P5), and 6171092 bp (P6). The average molecular mass per base pair (dsDNA) was defined as 660 g/mol.
Specificity of qPCR assays
In addition to 26 Pseudomonas strains, belonging to the seven target species (plus five very closely related species), 49 Pseudomonas strains from 45 non-target species and isolates of 40 other bacterial species were selected in order to check the specificity of the qPCR assays (Supplementary Table S1). Selection of strains was based on their relevance in raw milk and their phylogenetic proximity to the target species. Strains were grown on TS-agar plates for 24 h, and DNA was extracted using the QIAamp DNA Mini Kit (Qiagen N.V., Hilden, Germany) according to manufacturer’s specifications. DNA concentration was then measured via Qubit 2.0 Fluorometer (Invitrogen AG, Carlsbad, CA, USA) and adjusted to 1–2 ng/μl. One microliter DNA was used as template for the two triplex qPCR assays in a 10 μl reaction volume leading to a final DNA concentration of 0.1–0.2 ng/μl.
Generation of standard curves in raw milk
For the correlation of Cq values and cell counts, three to nine target strains were selected per species-specific probe and all of them (in total 26 strains) for the universal probe (Supplementary Table S1 in bold). Strains were grown in TSB at 30 °C and 150 rpm for 16 h, before 2 ml of each overnight culture was centrifuged (1 min, 13000g, RT). The supernatant was discarded, and the pellet resuspended in 10 ml of fresh raw milk. Afterward, a fivefold dilution series (1:51 to 1:59) of this sample was prepared with fresh raw milk. The dilution steps 1:52, 1:53, 1:55, 1:57, and 1:59 were selected for cell count determination by plating and DNA extraction using the DNeasy® PowerFood® Microbial Kit (Qiagen N.V., Hilden, Germany) with the above-listed protocol. Extracted, bacterial DNA of each strain was then applied as template (2 μl in 10 μl reaction volume) in the two triplex qPCR assays, and Cq values were determined of all samples in two technical replicates. For the rpoB probe (P6), the dilution step 1:52 was not considered due to the high amount of target DNA. In parallel, Pseudomonas counts of all sample dilutions were quantified on CFC-agar plates. The Pseudomonas count of untreated raw milk was determined (1.6 × 103 cfu/ml) and subtracted from the counts of spiked raw milk samples. For standard curves, logarithmic cell counts per milliliter were plotted against the respective Cq values of identical samples.
Results
Setup of multiplex qPCR for species-specific and total Pseudomonas detection
Besides the quantification of total pseudomonads, the novel qPCR assay was developed to specifically detect seven milk-relevant Pseudomonas species, namely P. proteolytica, P. gessardii, P. lactis, P. fluorescens, P. protegens, P. lundensis, and P. fragi. As target genes, aprX was chosen for species-specific detection, and the conserved rpoB gene for the quantification of total Pseudomonas counts.
In order to check the suitability of the selected target genes and to determine the number of probes and primers needed, phylogenetic analyses using aprX and rpoB sequences of 61 Pseudomonas strains, assigned to 46 different species, were performed. The overall topology of the phylogenetic aprX tree was similar to the one based on rpoB sequences (Fig. 1 and Supplementary Fig. S1), considering the classification of strains into the previously described Pseudomonas subgroups (Gomila et al. 2015; Peix et al. 2018; Maier et al. 2020) and the distribution of species within these groups. However, the aprX sequences were more variable and discriminative (66.0–100.0% sequence similarity range), thus enabling a higher resolution than the conserved rpoB gene sequences (85.1–100.0% sequence similarity range), which served for the design of a genus-specific probe and respective primers. With respect to the seven chosen target species, the aprX tree exhibited a distribution of the 18 representative strains in four Pseudomonas subgroups, namely P. fluorescens, P. gessardii, P. fragi, and P. chlororaphis. Sequences of isolates from P. lactis and P. fluorescens, both belonging to the P. fluorescens subgroup, as well as the ones from P. proteolytica and P. gessardii, being part of the P. gessardii subgroup, showed a very high inter-species sequence similarity of at least 91.9 and 94.1%, respectively. For these very closely related species, the design of a single hydrolysis probe and primer pair, targeting the aprX sequences from members of both species, was possible. In contrast, the aprX sequences of P. lundensis and P. fragi strains, all belonging to the P. fragi subgroup, differed largely (maximum inter-species sequence similarity of 81.1%), and therefore, separate probes and primers were created for each species. Also, for P. protegens strains, being located in the P. chlororaphis subgroup, the design of an additional probe plus primers was necessary. Consequently, a total of five hydrolysis probes (P1–P5) and primer pairs were generated to detect all seven target species. Besides the 18 strains of the target species, eight very closely related isolates were taken into account for probe and primer design (Supplementary Table S2), as they were shown to be frequently present in raw milk and exhibit comparable proteolytic characteristics (von Neubeck et al. 2015; Maier et al. 2020). According to our phylogenomic study, these strains do not belong to species with validly described names (Maier et al. 2020) and will be referred to as “P. lactis-like,” “P. gessardii-like,” “P. lundensis-like,” and “P. protegens-like” in the following.
Maximum likelihood phylogeny of the aprX gene based on 1,296 positions in the multiple sequence alignment of 61 distinct Pseudomonas strains. Molecular evolution was inferred by the GTR+G+I model, and the tree was outgroup-rooted (P. aeruginosa WS 5022). Branches with high bootstrap support (≥ 70% of 500 replicates) are marked with blue circles. Target strains of the species-specific probes are highlighted in blue (P1), orange (P2), brown (P3), green (P4), and red (P5). Strains were assigned to 13 monophyletic groups, whose names are listed in bold to the right of the tree
The six hydrolysis probes and respective primer pairs were split into two triplex qPCR reactions, whose compositions are summarized in Table 1. Assay 1 comprised three probes (P1–P3) and primer pairs to quantify common and highly proteolytic species, namely P. proteolytica, P. gessardii and P. gessardii-like species (P1); P. fluorescens, P. lactis and P. lactis-like species (P2); as well as P. protegens and P. protegens-like species (P3). Assay 2 contained two probes (P4 and P5) and respective primers detecting P. fragi (P4), and P. lundensis plus P. lundensis-like species (P5), which are less proteolytic, but abundant in raw milk samples. Moreover, assay 2 was complemented with the universal Pseudomonas primers and probe P6 for quantification of total Pseudomonas counts. For all designed primers and probes, the optimal concentrations were specified separately by singleplex qPCR (Table 1), and an optimal annealing temperature of 61 °C was determined by gradient qPCR (data not shown).
Efficiency and linearity of single- and multiplex qPCR
For determination of the amplification efficiencies, six defined DNA pools of target and non-target DNA were produced (Supplementary Table S3) and various dilutions thereof were applied as templates in qPCR. After testing each probe-primer combination separately in a singleplex assay, three probes plus primers were combined in the triplex approach. Therefore, DNA of two to six different target strains was applied for each of the five species-specific probes (P1–P5), and DNA of all 18 target strains was employed for the universal Pseudomonas probe (P6). Averaged Cq values were calculated for each hydrolysis probe with its primer pair (Table 2). Thereby, linear correlations between Cq values and DNA concentrations were observed for all six probes in singleplex and multiplex qPCR, yielding high correlation coefficient (R2) values of > 0.975 and PCR amplification efficiencies (E) of 85–97%. Since mean Cq values of singleplex and multiplex qPCRs were highly comparable (Table 2), possible interactions between the different probes and primer pairs in the multiplex reactions do not adversely affect target detection or amplification.
For the five species-specific probes (P1–P5), target DNA amounts from 2 to 0.0002 ng/μl (final concentrations in 10 μl reaction volume) and for the universal Pseudomonas probe P6 from 10 to 0.001 ng/μl were detectable, demonstrating a wide linear dynamic range over 4 log-steps. For qPCR with P1–P5, calculated minimal aprX gene copy numbers lay between 268 and 356. Using the universal Pseudomonas probe (P6), a minimum of approx. 1.5 × 103 rpoB gene copies was detectable with 0.001 ng/μl target DNA. Since no greater Cq value than 32.5 was received for all probes when applying the lowest target DNA amounts, a cut-off value of 33 was defined for further experiments, and higher Cq values were considered unquantifiable.
Specificity of the multiplex qPCR assay
To verify the specificity of the assay, 1–2 ng/μl genomic DNA of 75 Pseudomonas strains (target and non-target strains), assigned to 57 different species, and of 40 other bacterial species was applied as template in the qPCR assays. For strain selection, Pseudomonas isolates of the target species and their closest relatives, as well as representatives of the whole genus, were considered. Other bacterial species were chosen due to their phylogenetic proximity to the genus Pseudomonas and/or their relevance in milk and milk products.
Using the five species-specific probes (P1–P5) targeting aprX, all 18 strains of the defined seven target species were detected successfully, yielding Cq values from 18.85 to 22.13 (Table 3). Moreover, the eight very closely related isolates, which belong to P. gessardii-like, P. lactis-like, P. lundensis-like, and P. protegens-like species, resulted in positive signals in the same range (Cq 19.59–21.27).
For the 50 non-target pseudomonads tested, no false-positive signals were received using P3, P4, and P5, demonstrating a very high specificity. However, P1 and P2, detecting multiple target species at once, showed few false-positive results (Supplementary Table S4). In case of P1, a very weak signal was measured with DNA of P. marginalis DSM 17967 (Cq 32.1). Regarding P2, detecting P. lactis and P. fluorescens, false-positive signals were obtained for four strains of the closely related species Pseudomonas haemolytica, Pseudomonas paralactis, Pseudomonas orientalis, and Pseudomonas synxantha (Cq 24.4–27.6). For the 40 other bacterial species tested, no cross-reactivity was observed using P1–P5, except a negligible signal for Streptococcus pyogenes DSM 2071 (Cq 32.91) (Supplementary Table S4), underlining the high specificity of the designed primers and probes.
Via the universal rpoB probe (P6), 74 out of 75 Pseudomonas strains tested were successfully detected, generating Cq values between 17.45 and 22.69 (Supplementary Table 3 and Supplementary Table S4). Only the signal received from DNA of Pseudomonas stutzeri WS 5018 was considerably weaker (Cq 27.13). When testing the 40 other bacterial species, very weak unspecific signals were obtained for 5 isolates with P6 (Supplementary Table S4). Among them, Pseudoalteromonas haloplanktis WS 5482 yielded the highest signal with a Cq of 29.61, while the others showed even higher Cq values ranging from 30.96 to 32.91.
Quantification of Pseudomonas via multiplex qPCR
In order to generate standard curves for quantification of cells, Cq values from multiplex qPCR were correlated with the corresponding Pseudomonas counts from cultivation. Therefore, 26 Pseudomonas target strains were chosen (Supplementary Table S1), and cells were serially diluted in fresh raw milk. From selected dilution steps, cell counts were determined by cultivation, and in parallel, DNA was extracted and used as template for the two triplex qPCR assays.
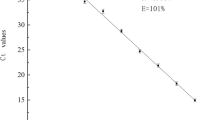
Linear correlation between Pseudomonas cell counts and Cq values was obtained for each of the six hydrolysis probes, revealing Cq values from 11.9 to 33.0 (Fig. 2). In agreement with the previously defined cut-off value, Cq values exceeding 33 were not detected. All standard curves yielded good efficiencies of 78–88% and high R2 values of 0.944 to 0.986. However, the R2 values of the probes P1, P2, and P6, detecting multiple species, were slightly lower than the ones from P3, P4, and P5, each targeting only a single species (Fig. 2, Table 3). The dynamic range of detection was found to be linear between ~ 103 and 107 cfu/ml for all probes, covering common Pseudomonas cell counts in raw milk. The lowest quantifiable cell amounts were also theoretically calculated for the stated Cq threshold of 33 via the standard curves, resulting in cell counts between 2 and 9 × 102 cfu/ml (P1: 561 cfu/ml; P2: 262 cfu/ml; P3: 556 cfu/ml; P4: 490 cfu/ml; P5: 359 cfu/ml; P6: 869 cfu/ml).
Standard curves of Cq values from multiplex qPCR using the six Pseudomonas probes P1–P6 (a–f), correlated with cell counts of artificially spiked raw milk samples. 26 representative target strains (9 strains for P1; 8 for P2; 3 for P3, P4 and P5; 26 for P6) were chosen, and cells were serially diluted in fresh raw milk (1:52–1:59). Cell counts of diluted samples were determined by cultivation, and in parallel, DNA of the milk samples was isolated and applied as templates in the two triplex qPCR assays. Averaged Cq values from two technical replicates of each strain were plotted against the respective Pseudomonas counts given in log cfu/ml. Regression equation, coefficient of determination (R2), and amplification efficiency (E) are given for each standard curve
Application of the qPCR assay to industrial raw milk samples
For assay validation, 60 independent raw milk samples from 13 different German dairies were analyzed. Determination of total and Pseudomonas cell counts by cultivation on TSA and CFC-agar, respectively, revealed great differences regarding the relative Pseudomonas amounts of samples (Supplementary Table S5).
For qPCR, bacterial DNA was isolated from raw milk samples and used as template in the two triplex assays. Based on the Cq values obtained, corresponding cell counts were calculated using the respective standard curve. Total Pseudomonas cell counts determined by qPCR via the universal Pseudomonas probe P6 ranged from 8.8 × 102 to 1.2 × 107 cfu/ml (Fig. 3). In two-thirds of the milk samples, cell counts from qPCR and from the cultivation approach did not differ more than 0.5 log, demonstrating a high concordance of the results. Almost one-third of samples showed a difference in cell amounts between 0.5 and 1 log, and only 2 from the 60 samples (no. 29 and 31) varied slightly more than 1 log (Fig. 3).
Comparison of Pseudomonas cell counts in 60 raw milk samples, determined by multiplex qPCR assay (black dots) and cultivation on CFC-agar (grey dots). For qPCR, DNA was isolated from each raw milk and applied as template in the assay using the universal Pseudomonas probe P6. Cell counts were calculated from received Cq values by linear regression analysis of a standard curve. Deviations between cell counts obtained with multiplex qPCR assay and plating are indicated by grey lines
Regarding the species composition, target species of the five species-specific probes (P1–P5) were detected by qPCRs in all but three raw milk samples tested, while non-target pseudomonads were detected in 53% of the samples (Fig. 4). Remarkably, the occurrence and proportion of each target species differed strongly among the raw milk samples. For each of the five species-specific probes, at least one milk sample contained exclusively the respective target species, confirming the usefulness of the chosen targets (Fig. 4). In terms of frequency and distribution, the species P. lundensis and/or P. lundensis-like (targeted by P5) were the most common species, being present in 80% of the milk samples and constituting the largest proportion of the Pseudomonas population in 27% of the milk samples. The target species of P1, namely P. proteolytica, P. gessardii and/or P. gessardii-like, were similarly frequent, being identified in 73% of the samples and predominant in 23% of the samples. The target species of P2 (P. fluorescens, P. lactis and/or P. lactis-like), P4 (P. fragi), and P3 (P. protegens and/or P. protegens-like) were detected in 60%, 42%, and 7% of the milk samples and accounted for the major share of pseudomonads in only 7%, 8%, and 2% of the samples, respectively. Finally, non-target pseudomonads presented the largest proportion in 33% of the raw milk samples (Fig. 4).
Distribution of different target species and non-target pseudomonads in 60 raw milk samples, determined by qPCR using five species-specific probes (P1–P5) and one universal Pseudomonas probe (P6). Cell counts of the target species and total Pseudomonas counts were calculated for each sample via standard curves and Cq values of the respective probes. Results from P6, representing the total count, were defined as 100%, and proportions of target strains accordingly determined. The proportion of non-target pseudomonads was defined by subtracting the sum of all target species (P1–P5) from the total Pseudomonas counts (P6)
In general, no correlation between total Pseudomonas cell counts and distribution of certain target species was observed in the analyzed raw milk samples; however, samples with Pseudomonas counts < 104 cfu/ml tended to comprise more different species than samples with higher cell counts.
Discussion
For the food industry, sensitive and rapid detection methods are crucial to perform a risk assessment and ensure the safety and quality of its products. In recent years, the development of multiplex qPCR assays to detect specific microorganisms in various food matrices has increased rapidly. Utilizing several probes with diverse fluorophores attached, multiplex qPCR enables the co-amplification and differentiation of multiple targets in a single reaction, presenting a cost- and time-saving alternative to singleplex qPCR or cultivation-dependent methods. So far, the majority of these applications allow the identification of foodborne pathogens. For example, multiplex qPCR assays have been developed for the detection of Salmonella spp., Bacillus cereus, Listeria monocytogenes, Staphylococcus aureus, Escherichia coli, Campylobacter spp., or Vibrio spp. in various foods (Hong et al. 2007; Tebbs et al. 2011; Forghani et al. 2016; Liu et al. 2017; Heymans et al. 2018; Parichehr et al. 2019). In addition, several applications deal with probiotic or beneficial organisms, such as yeasts, Acetobacter spp., or different lactic acid bacteria (LAB) in kefir or starter cultures in cheese production (Bottari et al. 2013; Nejati et al. 2020). Moreover, the detection of food-spoiling bacteria, e.g., Clostridium spp. in milk and meats or Bacillus spp. and Paenibacillus spp. in potato salad and milk, has been carried out by multiplex qPCR assays in previous studies (Morandi et al. 2015; Dorn-In et al. 2018; Nakanojp 2020).
For pseudomonads, two non-quantitative multiplex PCR approaches have been performed for different Pseudomonas species in meat (Ercolini et al. 2007) and of P. fluorescens strains with a biofilm-forming ability (Xu et al. 2017). However, until now, no qPCR assay has been developed for the simultaneous quantification of various milk-spoiling Pseudomonas species in raw milk, which would be very useful for quality assessment in the milk industry. The two triplex assays of this study resulted in the successful enumeration of total Pseudomonas counts as well as seven prevalent Pseudomonas species in raw milk, enabling discrimination of high and low peptidase producers. Regarding sensitivity, the assays exhibited a linear detection range of approx. 103–107 cfu/ml with lowest quantifiable cell numbers of 2 × 102–2 × 103 cfu/ml, depending on the TaqMan probe. These results are similar to the detection or quantification limits of other developed qPCR assays enumerating bacteria in spiked milk samples, e.g., Paenibacillus spp. and Bacillus spp. (Nakanojp 2020); E. coli and Salmonella spp. (Zhou et al. 2017); and S. aureus, L. monocytogenes, and Salmonella spp. (Ding et al. 2017). For some qPCR assays identifying foodborne pathogens in dairy products, lower detection limits of < 102 cfu/ml are required and obtained mostly via time-consuming enrichment steps or other sample pre-treatments (Forghani et al. 2016; Heymans et al. 2018; Parichehr et al. 2019). However, as previous studies revealed that average Pseudomonas cell counts in raw milk range from 102 to 105 cfu/ml (Leriche and Fayolle 2012; von Neubeck et al. 2015; Skeie et al. 2019), a higher sensitivity regarding the detection limit of our assay is neither necessary nor beneficial for its application.
When tested using 115 target- and non-target strains, our qPCR assay showed a high level of specificity. Only few false-positive signals were obtained for two out of the five species-specific hydrolysis probes, namely for the multi-target P1 and P2. For P2, this can be explained due to the close phylogenetic proximity of the target species P. lactis and P. fluorescens to the isolates causing false-positive signals (P. haemolytica DSM 108987, P. paralactis DSM 29164, P. orientalis DSM 17489, and P. synxantha DSM 18928). As all these strains were shown to be proteolytic, though less abundant in raw milk (von Neubeck et al. 2015; Maier et al. 2020), the signals are negligible or may even contribute to the detection of proteolytic pseudomonads. Moreover, the universal Pseudomonas probe (P6) was shown to be highly specific, detecting all 75 tested Pseudomonas strains. Among them, all tested isolates from the 15 species that were previously defined as milk relevant were found (von Neubeck et al. 2015; Caldera et al. 2016; Maier et al. 2020). When 40 non-pseudomonads were tested, the universal probe resulted in five very weak false-positive signals, the highest from DNA of P. haloplanktis WS 5482 (Cq 29.6). This psychrophilic marine bacterium has occasionally been isolated from cheese rind, but plays no role in the microbiota of raw milk (Feurer et al. 2004; Quigley et al. 2011; Almeida et al. 2014).
With respect to the enumeration of total Pseudomonas counts using P6, the results were in good agreement with cell counts received from cultivation. For the majority of samples, Pseudomonas cell counts quantified on selective agar were slightly higher than calculated cell counts via qPCR. This could be due to the growth of some members of Enterobacteriaceae or Acinetobacter on CFC-agar (Flint and Hartley 1996; Salvat et al. 1997), which are known to be frequently present in raw milk (Hantsis-Zacharov and Halpern 2007; Baur et al. 2015; von Neubeck et al. 2015; Li et al. 2018; Ribeiro Junior et al. 2018; Breitenwieser et al. 2020). Therefore, the determination of total Pseudomonas by our qPCR assay presents a highly specific and faster (3 h versus 2–3 days) alternative to the quantification of total counts by cultivation.
Remarkably, when 60 independent raw milk samples from 13 different dairies were analyzed for assay validation, unique compositions of the seven target species and non-target pseudomonads were detected for all samples. Thereby, P. lundensis and P. lundensis-like species (P5) were found most frequently (in 80% of the samples), closely followed by P. proteolytica, P. gessardii and P. gessardii-like species (P1). Members of P. lactis and P. fluorescens (P2) and P. fragi (P4) were also rather common (present in 60% and 42% of samples, respectively), while strains of P. protegens (P3) were relatively rare. Previous studies identifying the Pseudomonas population of raw milk or dairy products revealed the same predominant species, namely P. lundensis, P. proteolytica, P. gessardii, P. fragi, and P. fluorescens. In contrast, representatives of P. protegens were less common (Marchand et al. 2009a; De Jonghe et al. 2011; von Neubeck et al. 2015; Caldera et al. 2016). Besides, in all of these studies, other isolates from partly unclassified Pseudomonas species were identified, which is also consistent with our results revealing the presence of non-target species in about half of the samples tested.
Since it was shown that the composition of the Pseudomonas population varies greatly in raw milk samples, the proportions of highly, middle, and low proteolytic isolates are strongly different, too. Here, our two triplex qPCR assays offer a very useful tool to quantify and simultaneously distinguish between the most common raw milk species, possessing different proteolytic potentials. As triplex assay 1 detects specifically highly proteolytic Pseudomonas species (e.g., P. proteolytica, P. gessardii, or P. lactis) and assay 2 species with weaker peptidase activities (e.g., P. fragi and P. lundensis), they are well suited to estimate the spoilage potential of raw milk. However, for a more accurate risk assessment, future work is needed in order to determine the exact peptidase concentrations causing negative effects in milk. A previous study revealed product defects of UHT milk that was produced from raw milk contaminated with different Pseudomonas species, at peptidase activities of ≥ 0.03 pkat/ml (Stoeckel et al. 2016a). Correlations between AprX amounts and the required cell numbers of high as well as of low proteolytic Pseudomonas species are necessary for an informed definition of threshold CFU values, which indicate the probability of product spoilage.
In summary, the novel multiplex qPCR assay provides an accurate and rapid technique to quantify the total Pseudomonas counts in raw milk and to distinguish between the most prevalent Pseudomonas species with different proteolytic potentials. Thereby, it presents a powerful tool for the dairy industry to predict the spoilage risk and shelf life of raw milk samples at an early stage in order to decide on further processing, e.g., towards UHT or fresh milk products.
Data availability
All data generated during this study are included in the paper or in the electronic supplementary material. Strains and additional raw data are available from the authors upon request.
References
Almeida M, Hebert A, Abraham AL, Rasmussen S, Monnet C, Pons N, Delbes C, Loux V, Batto JM, Leonard P, Kennedy S, Ehrlich SD, Pop M, Montel MC, Irlinger F, Renault P (2014) Construction of a dairy microbial genome catalog opens new perspectives for the metagenomic analysis of dairy fermented products. BMC Genomics 15:1101. https://doi.org/10.1186/1471-2164-15-1101
Bagliniere F, Tanguy G, Jardin J, Mateos A, Briard V, Rousseau F, Robert B, Beaucher E, Humbert G, Dary A, Gaillard JL, Amiel C, Gaucheron F (2012) Quantitative and qualitative variability of the caseinolytic potential of different strains of Pseudomonas fluorescens: implications for the stability of casein micelles of UHT milks during their storage. Food Chem 135(4):2593–2603. https://doi.org/10.1016/j.foodchem.2012.06.099
Baur C, Krewinkel M, Kranz B, von Neubeck M, Wenning M, Scherer S, Stoeckel M, Hinrichs J, Stressler T, Fischer L (2015) Quantification of the proteolytic and lipolytic activity of microorganisms isolated from raw milk. Int Dairy J 49:23–29. https://doi.org/10.1016/j.idairyj.2015.04.005
Birkeland SE, Stepaniak L, Sorhaug T (1985) Quantitative studies of heat-stable proteinase from Pseudomonas fluorescens P1 by the enzyme-linked immunosorbent assay. Appl Environ Microbiol 49(2):382–387. https://doi.org/10.1128/AEM.49.2.382-387.1985
Bottari B, Agrimonti C, Gatti M, Neviani E, Marmiroli N (2013) Development of a multiplex real time PCR to detect thermophilic lactic acid bacteria in natural whey starters. Int J Food Microbiol 160(3):290–297. https://doi.org/10.1016/j.ijfoodmicro.2012.10.011
Breitenwieser F, Doll EV, Clavel T, Scherer S, Wenning M (2020) Complementary use of cultivation and high-throughput amplicon sequencing reveals high biodiversity within raw milk microbiota. Front Microbiol 11:1557. https://doi.org/10.3389/fmicb.2020.01557
Caldera L, Franzetti L, Van Coillie E, De Vos P, Stragier P, De Block J, Heyndrickx M (2016) Identification, enzymatic spoilage characterization and proteolytic activity quantification of Pseudomonas spp. isolated from different foods. Food Microbiol 54:142–153. https://doi.org/10.1016/j.fm.2015.10.004
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: Architecture and applications. BMC Bioinformatics 10:421. https://doi.org/10.1186/1471-2105-10-421
Clements RS, Wyatt DM, Symons MH, Ewings KN (1990) Inhibition enzyme-linked immunosorbent assay for detection of Pseudomonas fluorescens proteases in ultrahigh-temperature-treated milk. Appl Environ Microbiol 56(4):1188–1190. https://doi.org/10.1128/AEM.56.4.1188-1190.1990
De Jonghe V, Coorevits A, Van Hoorde K, Messens W, Van Landschoot A, De Vos P, Heyndrickx M (2011) Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl Environ Microbiol 77(2):460–470. https://doi.org/10.1128/AEM.00521-10
Ding T, Suo Y, Zhang Z, Liu D, Ye X, Chen S, Zhao Y (2017) A multiplex RT-PCR assay for S. aureus, L. monocytogenes, and Salmonella spp. detection in raw milk with pre-enrichment. Front Microbiol 8:989. https://doi.org/10.3389/fmicb.2017.00989
Dorn-In S, Schwaiger K, Springer C, Barta L, Ulrich S, Gareis M (2018) Development of a multiplex qPCR for the species identification of Clostridium estertheticum, C. frigoriphilum, C. bowmanii and C. tagluense-like from blown pack spoilage (BPS) meats and from wild boars. Int J Food Microbiol 286:162–169. https://doi.org/10.1016/j.ijfoodmicro.2018.08.020
Dufour D, Nicodeme M, Perrin C, Driou A, Brusseaux E, Humbert G, Gaillard JL, Dary A (2008) Molecular typing of industrial strains of Pseudomonas spp. isolated from milk and genetical and biochemical characterization of an extracellular protease produced by one of them. Int J Food Microbiol 125(2):188–196. https://doi.org/10.1016/j.ijfoodmicro.2008.04.004
Duong F, Bonnet E, Geli V, Lazdunski A, Murgier M, Filloux A (2001) The AprX protein of Pseudomonas aeruginosa: a new substrate for the Apr type I secretion system. Gene 262(1-2):147–153. https://doi.org/10.1016/s0378-1119(00)00541-2
Ercolini D, Russo F, Blaiotta G, Pepe O, Mauriello G, Villani F (2007) Simultaneous detection of Pseudomonas fragi, P. lundensis, and P. putida from meat by use of a multiplex PCR assay targeting the carA gene. Appl Environ Microbiol 73(7):2354–2359. https://doi.org/10.1128/AEM.02603-06
Feurer C, Irlinger F, Spinnler HE, Glaser P, Vallaeys T (2004) Assessment of the rind microbial diversity in a farmhouse-produced vs a pasteurized industrially produced soft red-smear cheese using both cultivation and rDNA-based methods. J Appl Microbiol 97(3):546–556. https://doi.org/10.1111/j.1365-2672.2004.02333.x
Flint S, Hartley N (1996) A modified selective medium for the detection of Pseudomonas species that cause spoilage of milk and dairy products. Int Dairy J 6(2):223–230. https://doi.org/10.1016/0958-6946(95)00007-0
Forghani F, Wei S, Oh DH (2016) A rapid multiplex real-time PCR high-resolution melt curve assay for the simultaneous detection of Bacillus cereus, Listeria monocytogenes, and Staphylococcus aureus in food. J Food Prot 79(5):810–815. https://doi.org/10.4315/0362-028X.JFP-15-428
Glück C, Rentschler E, Krewinkel M, Merz M, von Neubeck M, Wenning M, Scherer S, Stoeckel M, Hinrichs J, Stressler T, Fischer L (2016) Thermostability of peptidases secreted by microorganisms associated with raw milk. Int Dairy J 56:186–197. https://doi.org/10.1016/j.idairyj.2016.01.025
Gomila M, Pena A, Mulet M, Lalucat J, Garcia-Valdes E (2015) Phylogenomics and systematics in Pseudomonas. Front Microbiol 6:214. https://doi.org/10.3389/fmicb.2015.00214
Hantsis-Zacharov E, Halpern M (2007) Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl Environ Microbiol 73(22):7162–7168. https://doi.org/10.1128/AEM.00866-07
Heymans R, Vila A, van Heerwaarden CAM, Jansen CCC, Castelijn GAA, van der Voort M, Biesta-Peters EG (2018) Rapid detection and differentiation of Salmonella species, Salmonella Typhimurium and Salmonella Enteritidis by multiplex quantitative PCR. PLoS One 13(10):e0206316. https://doi.org/10.1371/journal.pone.0206316
Hong J, Jung WK, Kim JM, Kim SH, Koo HC, Ser J, Park YH (2007) Quantification and differentiation of Campylobacter jejuni and Campylobacter coli in raw chicken meats using a real-time PCR method. J Food Prot 70(9):2015–2022. https://doi.org/10.4315/0362-028x-70.9.2015
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. https://doi.org/10.1186/1471-2105-11-119
Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35(Web Server issue):W43–W46. https://doi.org/10.1093/nar/gkm234
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Lafarge V, Ogier JC, Girard V, Maladen V, Leveau JY, Gruss A, Delacroix-Buchet A (2004) Raw cow milk bacterial population shifts attributable to refrigeration. Appl Environ Microbiol 70(9):5644–5650. https://doi.org/10.1128/aem.70.9.5644-5650.2004
Leriche F, Fayolle K (2012) No seasonal effect on culturable pseudomonads in fresh milks from cattle herds. J Dairy Sci 95(5):2299–2306. https://doi.org/10.3168/jds.2011-4785
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47(W1):W256–W259. https://doi.org/10.1093/nar/gkz239
Li N, Wang Y, You C, Ren J, Chen W, Zheng H, Liu Z (2018) Variation in raw milk microbiota throughout 12 months and the impact of weather conditions. Sci Rep 8(1):2371. https://doi.org/10.1038/s41598-018-20862-8
Liu KC, Jinneman KC, Neal-McKinney J, Wu WH, Rice DH (2017) Simultaneous identification of Campylobacter jejuni, Campylobacter coli, and Campylobacter lari with SmartCycler-based multiplex quantitative polymerase chain reaction. Foodborne Pathog Dis 14(7):371–378. https://doi.org/10.1089/fpd.2016.2245
Machado SG, Bazzolli DMS, Vanetti MCD (2013) Development of a PCR method for detecting proteolytic psychrotrophic bacteria in raw milk. Int Dairy J 29(1):8–14. https://doi.org/10.1016/j.idairyj.2012.09.007
Machado SG, da Silva FL, Bazzolli DM, Heyndrickx M, Costa PM, Vanetti MC (2015) Pseudomonas spp. and Serratia liquefaciens as predominant spoilers in cold raw milk. J Food Sci 80(8):M1842–M1849. https://doi.org/10.1111/1750-3841.12957
Maier C, Huptas C, von Neubeck M, Scherer S, Wenning M, Lücking G (2020) Genetic organization of the aprX-lipA2 operon affects the proteolytic potential of Pseudomonas species in milk. Front Microbiol 11:1190. https://doi.org/10.3389/fmicb.2020.01190
Marchand S, Heylen K, Messens W, Coudijzer K, De Vos P, Dewettinck K, Herman L, De Block J, Heyndrickx M (2009a) Seasonal influence on heat-resistant proteolytic capacity of Pseudomonas lundensis and Pseudomonas fragi, predominant milk spoilers isolated from Belgian raw milk samples. Environ Microbiol 11(2):467–482. https://doi.org/10.1111/j.1462-2920.2008.01785.x
Marchand S, Vandriesche G, Coorevits A, Coudijzer K, De Jonghe V, Dewettinck K, De Vos P, Devreese B, Heyndrickx M, De Block J (2009b) Heterogeneity of heat-resistant proteases from milk Pseudomonas species. Int J Food Microbiol 133(1-2):68–77. https://doi.org/10.1016/j.ijfoodmicro.2009.04.027
Marchand S, Duquenne B, Heyndrickx M, Coudijzer K, De Block J (2017) Destabilization and off-flavors generated by Pseudomonas proteases during or after UHT-processing of milk. Int J Food Contam 4(1). https://doi.org/10.1186/s40550-016-0047-1
Martins ML, de Araujo EF, Mantovani HC, Moraes CA, Vanetti MC (2005) Detection of the apr gene in proteolytic psychrotrophic bacteria isolated from refrigerated raw milk. Int J Food Microbiol 102(2):203–211. https://doi.org/10.1016/j.ijfoodmicro.2004.12.016
Matéos A, Guyard-Nicodème M, Baglinière F, Jardin J, Gaucheron F, Dary A, Humbert G, Gaillard JL (2015) Proteolysis of milk proteins by AprX, an extracellular protease identified in Pseudomonas LBSA1 isolated from bulk raw milk, and implications for the stability of UHT milk. Int Dairy J 49:78–88. https://doi.org/10.1016/j.idairyj.2015.04.008
Matta H, Punj V, Kanwar SS (1997) An immuno-dot blot assay for detection of thermostable protease from Pseudomonas sp. AFT-36 of dairy origin. Lett Appl Microbiol 25(4):300–302. https://doi.org/10.1046/j.1472-765x.1997.00228.x
McKellar RC (1981) Development of off-flavors in ultra-high temperature and pasteurized milk as a function of proteolysis. J Dairy Sci 64:2138–2145. https://doi.org/10.3168/jds.S0022-0302(81)82820-2
Morandi S, Cremonesi P, Silvetti T, Castiglioni B, Brasca M (2015) Development of a triplex real-time PCR assay for the simultaneous detection of Clostridium beijerinckii, Clostridium sporogenes and Clostridium tyrobutyricum in milk. Anaerobe 34:44–49. https://doi.org/10.1016/j.anaerobe.2015.04.005
Nakanojp M (2020) Development of a multiplex real-time PCR assay for the identification and quantification of group-specific Bacillus spp. and the genus Paenibacillus. Int J Food Microbiol 323:108573. https://doi.org/10.1016/j.ijfoodmicro.2020.108573
Nejati F, Junne S, Kurreck J, Neubauer P (2020) Quantification of major bacteria and yeast species in kefir consortia by multiplex TaqMan qPCR. Front Microbiol 11:1291. https://doi.org/10.3389/fmicb.2020.01291
Parichehr M, Mohammad K, Abbas D, Mehdi K (2019) Developing a multiplex real-time PCR with a new pre-enrichment to simultaneously detect four foodborne bacteria in milk. Future Microbiol 14:885–898. https://doi.org/10.2217/fmb-2019-0044
Peix A, Ramirez-Bahena MH, Velazquez E (2018) The current status on the taxonomy of Pseudomonas revisited: an update. Infect Genet Evol 57:106–116. https://doi.org/10.1016/j.meegid.2017.10.026
Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD (2011) Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int J Food Microbiol 150(2-3):81–94. https://doi.org/10.1016/j.ijfoodmicro.2011.08.001
Ribeiro Junior JC, de Oliveira AM, Silva FG, Tamanini R, de Oliveira ALM, Beloti V (2018) The main spoilage-related psychrotrophic bacteria in refrigerated raw milk. J Dairy Sci 101(1):75–83. https://doi.org/10.3168/jds.2017-13069
Salvat G, Rudelle S, Humbert F, Colin P, Lahellec C (1997) A selective medium for the rapid detection by an impedance technique of Pseudomonas spp. associated with poultry meat. J Appl Microbiol 83(4):456–463. https://doi.org/10.1046/j.1365-2672.1997.00256.x
Schokker EP, van Boekel MAJS (1997) Production, purification and partial characterization of the extracellular proteinase from Pseudomonas fluorescens 22F. Int Dairy J 7(4):265–271. https://doi.org/10.1016/s0958-6946(97)00008-3
Skeie SB, Haland M, Thorsen IM, Narvhus J, Porcellato D (2019) Bulk tank raw milk microbiota differs within and between farms: a moving goalpost challenging quality control. J Dairy Sci 102(3):1959–1971. https://doi.org/10.3168/jds.2017-14083
Sørhaug T, Stepaniak L (1997) Psychrotrophs and their enzymes in milk and dairy products: quality aspects. Trends Food Sci Technol 8(2):35–41. https://doi.org/10.1016/s0924-2244(97)01006-6
Staroscik A (2011-2020) Copy number calculator for realtime PCR. http://scienceprimer.com/copy-number-calculator-for-realtime-pcr. Accessed 27 June 2020
Stoeckel M, Lidolt M, Achberger V, Glück C, Krewinkel M, Stressler T, von Neubeck M, Wenning M, Scherer S, Fischer L, Hinrichs J (2016a) Growth of Pseudomonas weihenstephanensis, Pseudomonas proteolytica and Pseudomonas sp. in raw milk: impact of residual heat-stable enzyme activity on stability of UHT milk during shelf-life. Int Dairy J 59:20–28. https://doi.org/10.1016/j.idairyj.2016.02.045
Stoeckel M, Lidolt M, Stressler T, Fischer L, Wenning M, Hinrichs J (2016b) Heat stability of indigenous milk plasmin and proteases from Pseudomonas: a challenge in the production of ultra-high temperature milk products. Int Dairy J 61:250–261. https://doi.org/10.1016/j.idairyj.2016.06.009
Tebbs RS, Brzoska PM, Furtado MR, Petrauskene OV (2011) Design and validation of a novel multiplex real-time PCR assay for Vibrio pathogen detection. J Food Prot 74(6):939–948. https://doi.org/10.4315/0362-028X.JFP-10-511
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. https://doi.org/10.1093/nar/22.22.4673
von Neubeck M, Baur C, Krewinkel M, Stoeckel M, Kranz B, Stressler T, Fischer L, Hinrichs J, Scherer S, Wenning M (2015) Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int J Food Microbiol 211:57–65. https://doi.org/10.1016/j.ijfoodmicro.2015.07.001
von Neubeck M, Huptas C, Gluck C, Krewinkel M, Stoeckel M, Stressler T, Fischer L, Hinrichs J, Scherer S, Wenning M (2016) Pseudomonas helleri sp. nov. and Pseudomonas weihenstephanensis sp. nov., isolated from raw cow’s milk. Int J Syst Evol Microbiol 66(3):1163–1173. https://doi.org/10.1099/ijsem.0.000852
Woods RG, Burger M, Beven CA, Beacham IR (2001) The aprX-lipA operon of Pseudomonas fluorescens B52: a molecular analysis of metalloprotease and lipase production. Microbiology 147(Pt 2):345–354. https://doi.org/10.1099/00221287-147-2-345
Xu Y, Chen W, You C, Liu Z (2017) Development of a multiplex PCR assay for detection of Pseudomonas fluorescens with biofilm formation ability. J Food Sci 82(10):2337–2342. https://doi.org/10.1111/1750-3841.13845
Zhou B, Liang T, Zhan Z, Liu R, Li F, Xu H (2017) Rapid and simultaneous quantification of viable Escherichia coli O157:H7 and Salmonella spp. in milk through multiplex real-time PCR. J Dairy Sci 100(11):8804–8813. https://doi.org/10.3168/jds.2017-13362
Acknowledgements
We would like to thank Angela Felsl and Inge Celler for outstanding technical support.
Funding
Open Access funding enabled and organized by Projekt DEAL. Open Access funding is provided by Projekt DEAL (German DEAL agreement). This IGF Project of the FEI (no. AiF 18326 N) was supported via AiF within the program for promoting the Industrial Collective Research (IGF) of the German Ministry of Economic Affairs and Energy (BMWi), based on a resolution of the German Parliament.
Author information
Authors and Affiliations
Contributions
CM: performed research, analyzed data, and wrote manuscript; KH: conducted experiments and analyzed data; CH: prepared phylogenetic trees and wrote sections of material and methods; SS: discussed and revised manuscript; MW: designed study and revised manuscript; GL: planned study and wrote manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 1.15 mb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maier, C., Hofmann, K., Huptas, C. et al. Simultaneous quantification of the most common and proteolytic Pseudomonas species in raw milk by multiplex qPCR. Appl Microbiol Biotechnol 105, 1693–1708 (2021). https://doi.org/10.1007/s00253-021-11109-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11109-0