Regulation of CREB Phosphorylation in Nucleus Accumbens after Relief Conditioning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal
2.2. Apparatus
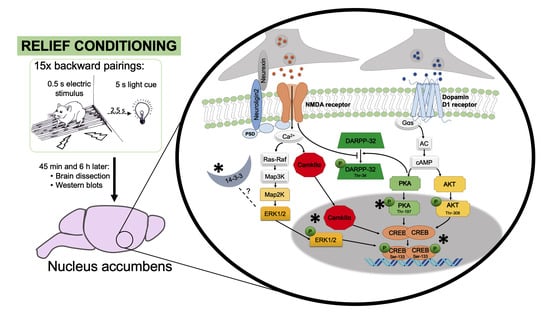
2.3. Behavioral Procedure
2.4. Experimental Design
2.5. Tissue Isolation
2.6. Protein Extraction (from Brain Samples) and Western Blot Analysis
2.7. Statistical Analysis
3. Results
3.1. Behavioral Analysis
3.2. Analysis of Protein Expression, Protein Phosphorylation, and Phosphorylation Ratios
3.2.1. Relief Learning Affects CREB Expression Time-Dependent and Brain Region-Specifically
3.2.2. Moderate Induction of Kinases Underlying NMDA Receptor Signaling Plays a Role in Relief Learning
3.2.3. Dopamine D1 Receptor Mediated PKA Activation Is Involved in CREB Phosphorylation after Relief Conditioning
3.2.4. 14-3-3 and Neuroligin2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gerber, B.; Yarali, A.; Diegelmann, S.; Wotjak, C.T.; Pauli, P.; Fendt, M. Pain-relief learning in flies, rats, and man: Basic research and applied perspectives. Learn. Mem. 2014, 21, 232–252. [Google Scholar] [CrossRef] [Green Version]
- Navratilova, E.; Atcherley, C.W.; Porreca, F. Brain Circuits Encoding Reward from Pain Relief. Trends Neurosci. 2015, 38, 741–750. [Google Scholar] [CrossRef] [Green Version]
- Leknes, S.; Lee, M.; Berna, C.; Andersson, J.; Tracey, I. Relief as a reward: Hedonic and neural responses to safety from pain. PLoS ONE 2011, 6, e17870. [Google Scholar] [CrossRef]
- Gerber, B.; König, C.; Fendt, M.; Andreatta, M.; Romanos, M.; Pauli, P.; Yarali, A. Timing-dependent valence reversal: A principle of reinforcement processing and its possible implications. Curr. Opin. Behav. Sci. 2019, 26, 114–120. [Google Scholar] [CrossRef]
- Navratilova, E.; Porreca, F. Reward and motivation in pain and pain relief. Nat. Neurosci. 2014, 17, 1304–1312. [Google Scholar] [CrossRef]
- Andreatta, M.; Fendt, M.; Mühlberger, A.; Wieser, M.J.; Imobersteg, S.; Yarali, A.; Gerber, B.; Pauli, P. Onset and offset of aversive events establish distinct memories requiring fear and reward networks. Learn. Mem. 2012, 19, 518–526. [Google Scholar] [CrossRef] [Green Version]
- Mayer, D.; Kahl, E.; Uzuneser, T.C.; Fendt, M. Role of the mesolimbic dopamine system in relief learning. Neuropsychopharmacology 2018, 43, 1651–1659. [Google Scholar] [CrossRef] [Green Version]
- Smith-Roe, S.L.; Kelley, A.E. Coincident activation of NMDA and dopamine D1 receptors within the nucleus accumbens core is required for appetitive instrumental learning. J. Neurosci. 2000, 20, 7737–7742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammadi, M.; Fendt, M. Relief learning is dependent on NMDA receptor activation in the nucleus accumbens. Br. J. Pharm. 2015, 172, 2419–2426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergado Acosta, J.R.; Kahl, E.; Kogias, G.; Uzuneser, T.C.; Fendt, M. Relief learning requires a coincident activation of dopamine D1 and NMDA receptors within the nucleus accumbens. Neuropharmacology 2017, 114, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Kandel, E.R. The molecular biology of memory: CAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 2012, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaldun, J.C.; Sprecher, S.G. Initiated by CREB: Resolving Gene Regulatory Programs in Learning and Memory: Switch in Cofactors and Transcription Regulators between Memory Consolidation and Maintenance Network. BioEssays 2019, 41, 1900045. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, J.; Lazarovici, P.; Quirion, R.; Zheng, W. cAMP Response Element-Binding Protein (CREB): A Possible Signaling Molecule Link in the Pathophysiology of Schizophrenia. Front. Mol. Neurosci. 2018, 11, 255. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, M.; Radulovic, J.; Spiess, J. Phosphorylated cAMP response element binding protein in the mouse brain after fear conditioning: Relationship to Fos production. Mol. Brain Res. 2001, 94, 15–24. [Google Scholar] [CrossRef]
- Heffner, T.G.; Hartman, J.A.; Seiden, L.S. A rapid method for the regional dissection of the rat brain. Pharmacol. Biochem. Behav. 1980, 13, 453–456. [Google Scholar] [CrossRef]
- Badin, J.; Herve, B. La Pr’Ecipitation Des Prot’Eines Par L’Amidoschwarz Et Son Application Au Microdosage Des Prot’Eines S’Eriques (Protein precipitation by amidoschwary and its application to the microdetermination of blood peoteins). Ann. Biol. Clin. (Paris) 1965, 23, 321–332. [Google Scholar] [PubMed]
- Thomas, G.M.; Huganir, R.L. MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 2004, 5, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Sheng, M.; McFadden, G.; Greenberg, M.E. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron 1990, 4, 571–582. [Google Scholar] [CrossRef]
- Tropea, T.F.; Kosofsky, B.E.; Rajadhyaksha, A.M. Enhanced CREB and DARPP-32 phosphorylation in the nucleus accumbens and CREB, ERK, and GluR1 phosphorylation in the dorsal hippocampus is associated with cocaine-conditioned place preference behavior. J. Neurochem. 2008, 106, 1780–1790. [Google Scholar] [CrossRef]
- Nairn, A.C.; Svenningsson, P.; Nishi, A.; Fisone, G.; Girault, J.A.; Greengard, P. The role of DARPP-32 in the actions of drugs of abuse. Neuropharmacology 2004, 47, 14–23. [Google Scholar] [CrossRef]
- Kähne, T.; Richter, S.; Kolodziej, A.; Smalla, K.H.; Pielot, R.; Engler, A.; Ohl, F.W.; Dieterich, D.C.; Seidenbecher, C.; Tischmeyer, W.; et al. Proteome rearrangements after auditory learning: High-resolution profiling of synapse-enriched protein fractions from mouse brain. J. Neurochem. 2016, 138, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Rescola, R.A. Pavlovian Conditioning and Its Proper Control Procedures. Psychol. Rev. 1967, 74, 71–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, A.J.; Kogan, J.H.; Frankland, P.W.; Kida, S. CREB and memory. Annu. Rev. Neurosci. 1998, 21, 127–148. [Google Scholar] [CrossRef] [Green Version]
- Barrot, M.; Olivier, J.D.A.; Perrotti, L.I.; DiLeone, R.J.; Berton, O.; Eisch, A.J.; Impey, S.; Storm, D.R.; Neve, R.L.; Yin, J.C.; et al. CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proc. Natl. Acad. Sci. USA 2002, 99, 11435–11440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega-Martínez, S. A new perspective on the role of the CREB family of transcription factors in memory consolidation via adult hippocampal neurogenesis. Front. Mol. Neurosci. 2015, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, K.; Karelina, K.; Obrietan, K. CREB: A multifaceted regulator of neuronal plasticity and protection. J. Neurochem. 2011, 116, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Dudai, Y.; Eisenberg, M. Rites of passage of the engram: Reconsolidation and the lingering consolidation hypothesis. Neuron 2004, 44, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Trifilieff, P.; Herry, C.; Vanhoutte, P.; Caboche, J.; Desmedt, A.; Riedel, G.; Mons, N.; Micheau, J. Foreground contextual fear memory consolidation requires two independent phases of hippocampal ERK/CREB activation. Learn. Mem. 2006, 13, 349–358. [Google Scholar] [CrossRef] [Green Version]
- Bito, H.; Deisseroth, K.; Tsien, R.W. CREB phosphorylation and dephosphorylation: A Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell 1996, 87, 1203–1214. [Google Scholar] [CrossRef] [Green Version]
- Ghiani, C.A.; Beltran-Parrazal, L.; Sforza, D.M.; Malvar, J.S.; Seksenyan, A.; Cole, R.; Smith, D.J.; Charles, A.; Ferchmin, P.A.; De Vellis, J. Genetic program of neuronal differentiation and growth induced by specific activation of NMDA receptors. Neurochem. Res. 2007, 32, 363–376. [Google Scholar] [CrossRef]
- Eagle, D.M.; Baunez, C. Is there an inhibitory-response-control system in the rat? Evidence from anatomical and pharmacological studies of behavioral inhibition. Neurosci. Biobehav. Rev. 2010, 34, 50–72. [Google Scholar] [CrossRef] [PubMed]
- Alberini, C.M. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 2009, 89, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, R.Y. Basic roles of key molecules connected with NMDAR signaling pathway on regulating learning and memory and synaptic plasticity. Mil. Med. Res. 2016, 3, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abel, T.; Nguyen, P.V. Chapter 6 Regulation of hippocampus-dependent memory by cyclic AMP-dependent protein kinase. Prog. Brain Res. 2008, 169, 97–115. [Google Scholar] [PubMed] [Green Version]
- Brami-Cherrier, K.; Valjent, E.; Garcia, M.; Pagès, C.; Hipskind, R.A.; Caboche, J. Dopamine induces a PI3-kinase-independent activation of Akt in striatal neurons: A new route to cAMP response element-binding protein phosphorylation. J. Neurosci. 2002, 22, 8911–8921. [Google Scholar] [CrossRef] [Green Version]
- Nishi, A.; Matamales, M.; Musante, V.; Valjent, E.; Kuroiwa, M.; Kitahara, Y.; Rebholz, H.; Greengard, P.; Girault, J.A.; Nairn, A.C. Glutamate counteracts dopamine/pka signaling via dephosphorylation of DARPP-32 Ser-97 and alteration of its cytonuclear distribution. J. Biol. Chem. 2017, 292, 1462–1476. [Google Scholar] [CrossRef] [Green Version]
- Nishi, A.; Bibb, J.A.; Snyder, G.L.; Higashi, H.; Nairn, A.C.; Greengard, P. Amplification of dopaminergic signaling by a positive feedback loop. Proc. Natl. Acad. Sci. USA 2000, 97, 12840–12845. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhou, Y. 14-3-3 Proteins in Glutamatergic Synapses. Neural Plast. 2018, 2018, 8407609. [Google Scholar] [CrossRef]
- Skoulakis, E.M.C.; Davis, R.L. 14-3-3 Proteins in neuronal development and function. Mol. Neurobiol. 1998, 16, 269–284. [Google Scholar] [CrossRef]
- Qiao, H.; Foote, M.; Graham, K.; Wu, Y.; Zhou, Y. 14-3-3 Proteins Are Required for Hippocampal Long-Term Potentiation and Associative Learning and Memory. J. Neurosci. 2014, 34, 4801–4808. [Google Scholar] [CrossRef]
- Wu, Y.-J.; Ko, B.-S.; Liou, J.-Y. 14-3-3. In Encyclopedia of Signaling Molecules; Springer: New York, NY, USA, 2018; pp. 1–11. [Google Scholar]
- Freed, E.; Symons, M.; Macdonald, S.G.; McCormick, F.; Ruggieri, R. Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science 1994, 265, 1713–1716. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Zhang, C.; Zhang, Q.; Sahin, O.; Wang, H.; Xu, J.; Xiao, Y.; Zhang, J.; Rehman, S.K.; Li, P.; et al. Upregulation of lactate dehydrogenase a by 14-3-3ζ leads to increased glycolysis critical for breast cancer initiation and progression. Oncotarget 2016, 7, 35270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Yu, S.; Fu, Y.; Li, X. Synaptic proteins and receptors defects in autism spectrum disorders. Front. Cell. Neurosci. 2014, 8, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bollen, E.; Puzzo, D.; Rutten, K.; Privitera, L.; De Vry, J.; Vanmierlo, T.; Kenis, G.; Palmeri, A.; D’Hooge, R.; Balschun, D.; et al. Improved long-term memory via enhancing cGMP-PKG signaling requires cAMP-PKA signaling. Neuropsychopharmacology 2014, 39, 2497–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goode, T.D.; Ressler, R.L.; Acca, G.M.; Miles, O.W.; Maren, S. Bed nucleus of the stria terminalis regulates fear to unpredictable threat signals. eLife 2019, 8, e46525. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.F.; Kandel, E.R.; Hawkins, R.D. Nitric oxide signaling contributes to late-phase LTP and CREB phosphorylation in the hippocampus. J. Neurosci. 1999, 19, 10250–10261. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soleimanpour, E.; Bergado Acosta, J.R.; Landgraf, P.; Mayer, D.; Dankert, E.; Dieterich, D.C.; Fendt, M. Regulation of CREB Phosphorylation in Nucleus Accumbens after Relief Conditioning. Cells 2021, 10, 238. https://doi.org/10.3390/cells10020238
Soleimanpour E, Bergado Acosta JR, Landgraf P, Mayer D, Dankert E, Dieterich DC, Fendt M. Regulation of CREB Phosphorylation in Nucleus Accumbens after Relief Conditioning. Cells. 2021; 10(2):238. https://doi.org/10.3390/cells10020238
Chicago/Turabian StyleSoleimanpour, Elaheh, Jorge R. Bergado Acosta, Peter Landgraf, Dana Mayer, Evelyn Dankert, Daniela C. Dieterich, and Markus Fendt. 2021. "Regulation of CREB Phosphorylation in Nucleus Accumbens after Relief Conditioning" Cells 10, no. 2: 238. https://doi.org/10.3390/cells10020238