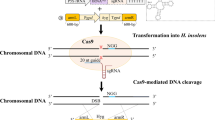
Abstract
Objective
To construct convenient CRISPR/Cas9-mediated genome editing systems in industrial enzyme-producing fungi Penicillium oxalicum and Trichoderma reesei.
Results
Employing the 5S rRNA promoter from Aspergillus niger for guide RNA expression, the β-glucosidase gene bgl2 in P. oxalicum was deleted using a donor DNA carrying 40-bp homology arms or a donor containing no selectable marker gene. Using a markerless donor DNA as editing template, precise replacement of a small region was achieved in the creA gene. In T. reesei, the A. niger 5S rRNA promoter was less efficient than that in P. oxalicum when used for gene editing. Using a native 5S rRNA promoter, stop codons were introduced into the lae1 coding region using a markerless donor DNA with an editing efficiency of 36.67%.
Conclusions
Efficient genome editing systems were developed in filamentous fungi P. oxalicum and T. reesei by using heterologous or native 5S rRNA promoters for guide RNA expression.




Similar content being viewed by others
References
Adnan M, Zheng W, Islam W, Arif M, Abubakar YS, Wang Z, Lu G (2017) Carbon catabolite repression in filamentous fungi. Int J Mol Sci 19:48. https://doi.org/10.3390/ijms19010048
Bischof RH, Ramoni J, Seiboth B (2016) Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb Cell Fact 15:106. https://doi.org/10.1186/s12934-016-0507-6
Chen M et al (2013) Promotion of extracellular lignocellulolytic enzymes production by restraining the intracellular beta-glucosidase in Penicillium decumbens. Bioresour Technol 137:33–40. https://doi.org/10.1016/j.biortech.2013.03.099
Cheng Z et al (2016) A novel acid-stable endo-polygalacturonase from Penicillium oxalicum CZ1028: purification, characterization, and application in the beverage industry. J Microbiol Biotechnol 26:989–998. https://doi.org/10.4014/jmb.1511.11045
Cullen D, Leong SA, Wilson LJ, Henner DJ (1987) Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene 57:21–26. https://doi.org/10.1016/0378-1119(87)90172-7
Deane EE, Whipps JM, Lynch JM, Peberdy JF (1999) Transformation of Trichoderma reesei with a constitutively expressed heterologous fungal chitinase gene. Enzyme Microb Tech 24:419–424. https://doi.org/10.1016/S0141-0229(98)00155-0
Deng H, Gao R, Liao X, Cai Y (2017) CRISPR system in filamentous fungi: current achievements and future directions. Gene 627:212–221. https://doi.org/10.1016/j.gene.2017.06.019
Fonseca LM, Parreiras LS, Murakami MT (2020) Rational engineering of the Trichoderma reesei RUT-C30 strain into an industrially relevant platform for cellulase production. Biotechnol Biofuels 13:93. https://doi.org/10.1186/s13068-020-01732-w
Gao L et al (2017) Combining manipulation of transcription factors and overexpression of the target genes to enhance lignocellulolytic enzyme production in Penicillium oxalicum. Biotechnol Biofuels 10:100. https://doi.org/10.1186/s13068-017-0783-3
Häkkinen M et al (2014) Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels 7:14. https://doi.org/10.1186/1754-6834-7-14
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98. https://doi.org/10.1021/bk-1999-0734.ch008
Hao Z, Su X (2019) Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex. BMC Biotechnol 19:2. https://doi.org/10.1186/s12896-018-0498-y
Katayama T, Nakamura H, Zhang Y, Pascal A, Fujii W, Maruyama JI (2019) Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae. Appl Environ Microbiol 85:e01896-e1918. https://doi.org/10.1128/AEM.01896-18
Krappmann S (2017) CRISPR-Cas9, the new kid on the block of fungal molecular biology. Med Mycol 55:16–23. https://doi.org/10.1093/mmy/myw097
Kun RS, Meng JL, Salazar-Cerezo S, Mäkelä MR, de Vries RP, Garrigues S (2020) CRISPR/Cas9 facilitates rapid generation of constitutive forms of transcription factors in Aspergillus niger through specific on-site genomic mutations resulting in increased saccharification of plant biomass. Enzyme Microb Tech 136:109508. https://doi.org/10.1016/j.enzmictec.2020.109508
Kwon MJ, Schütze T, Spohner S, Haefner S, Meyer V (2019) Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi. Fungal Biol Biotechnol 6:15. https://doi.org/10.1186/s40694-019-0079-4
Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E (2019) CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 47:W171–W174. https://doi.org/10.1093/nar/gkz365
Larkin MA et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Leynaud-Kieffer LMC, Curran SC, Kim I, Magnuson JK, Gladden JM, Baker SE, Simmons BA (2019) A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable. PLoS One 14:e0210243. https://doi.org/10.1371/journal.pone.0210243
Liu G et al (2013) Long-term strain improvements accumulate mutations in regulatory elements responsible for hyper-production of cellulolytic enzymes. Sci Rep 3:1569. https://doi.org/10.1038/srep01569
Liu G, Qin Y, Li Z, Qu Y (2013) Improving lignocellulolytic enzyme production with Penicillium: from strain screening to systems biology. Biofuels 4:523–534
Liu Q, Zhang Y, Li F, Li J, Sun W, Tian C (2019) Upgrading of efficient and scalable CRISPR-Cas-mediated technology for genetic engineering in thermophilic fungus Myceliophthora thermophila. Biotechnol Biofuels 12:293. https://doi.org/10.1186/s13068-019-1637-y
Liu R, Chen L, Jiang Y, Zhou Z, Zou G (2015) Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov 1:15007. https://doi.org/10.1038/celldisc.2015.7
Oakley BR, Rinehart JE, Mitchell BL, Oakley CE, Carmona C, Gray GL, May GS (1987) Cloning, mapping and molecular analysis of the pyrG (orotidine-5’-phosphate decarboxylase) gene of Aspergillus nidulans. Gene 61:385–399. https://doi.org/10.1016/0378-1119(87)90201-0
Penttilä M, Nevalainen H, Rättö M, Salminen E, Knowles J (1987) A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155–164. https://doi.org/10.1016/0378-1119(87)90110-7
Qin Y, Zheng K, Liu G, Chen M, Qu Y (2013) Improved cellulolytic efficacy in Penicilium decumbens via heterologous expression of Hypocrea jecorina endoglucanase II. Arch Biol Sci, Belgrade 65:305–314. https://doi.org/10.2298/ABS1301305Q
Rantasalo A, Vitikainen M, Paasikallio T, Jäntti J, Landowski CP, Mojzita D (2019) Novel genetic tools that enable highly pure protein production in Trichoderma reesei. Sci Rep 9:5032. https://doi.org/10.1038/s41598-019-41573-8
Ries LNA, Beattie SR, Espeso EA, Cramer RA, Goldman GH (2016) Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics 203:335–352. https://doi.org/10.1534/genetics.116.187872
Saini R, Saini JK, Adsul M, Patel AK, Mathur A, Tuli D, Singhania RR (2015) Enhanced cellulase production by Penicillium oxalicum for bio-ethanol application. Bioresour Technol 188:240–246. https://doi.org/10.1016/j.biortech.2015.01.048
Seiboth B et al (2012) The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol Microbiol 84:1150–1164. https://doi.org/10.1111/j.1365-2958.2012.08083.x
Shi T et al (2017) CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl Microbiol Biotechnol 101:7435–7443. https://doi.org/10.1007/s00253-017-8497-9
Shi T, Gao J, Wang W, Wang K, Xu G, Huang H, Ji X (2019) CRISPR/Cas9-based genome editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth Biol 8:445–454. https://doi.org/10.1021/acssynbio.8b00478
Szymanski M, Zielezinski A, Barciszewski J, Erdmann VA, Karlowski WM (2016) 5SRNAdb: an information resource for 5S ribosomal RNAs. Nucleic Acids Res 44:D180-183. https://doi.org/10.1093/nar/gkv1081
Vogel H (1956) A convenient growth medium for Neurospora (Medium N). Microb Genet Bull 13:42–43
Wang L et al (2018) Efficient CRISPR-Cas9 mediated multiplex genome editing in yeasts. Biotechnol Biofuels 11:277. https://doi.org/10.1186/s13068-018-1271-0
Wang L, Zhao S, Chen XX, Deng Q, Li C, Feng J (2018) Secretory overproduction of a raw starch-degrading glucoamylase in Penicillium oxalicum using strong promoter and signal peptide. Appl Microbiol Biotechnol 102:9291–9301. https://doi.org/10.1007/s00253-018-9307-8
Wu C et al (2020) A simple approach to mediate genome editing in the filamentous fungus Trichoderma reesei by CRISPR/Cas9-coupled in vivo gRNA transcription. Biotechnol Lett 42:1203–1210. https://doi.org/10.1007/s10529-020-02887-0
Zheng X, Zheng P, Zhang K, Cairns TC, Meyer V, Sun J, Ma Y (2019) 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger. ACS Synth Biol 8:1568–1574. https://doi.org/10.1021/acssynbio.7b00456
Zhong L, Qian Y, Dai M, Zhong Y (2016) Improvement of uracil auxotrophic transformation system in Trichoderma reesei QM9414 and overexpression of β-glucosidase. CIESC J 67:2510–2518. https://doi.org/10.11949/j.issn.0438-1157.20151654
Acknowledgements
We thank Xiaomei Zheng, Jibin Sun and Weifeng Liu for providing plasmids. This work was supported by National Key R&D Program of China Grant (2018YFA0900500), Major Basic Research Program of Shandong Provincial Natural Science Foundation (ZR2019ZD19), the Key Research and Development Project of Shandong Province (2019JZZY020223 and 2019JZZY020807), and the Young Scholars Program of Shandong University (YSPSDU) (to G. L.).
Supplementary Materials
Fig. S1 Plasmids containing Cas9 expression cassettes.
Fig. S2 The growth of P. oxalicum M12 and 9-10 on 2% (w/v) glucose agar plate.
Fig. S3 Diagnostic PCR analysis of P. oxalicum bgl2 deletion transformants.
Fig. S4 Diagnostic PCR analysis of P. oxalicum creA editing transformants.
Table S1 Sequences of protospacers in this study.
Table S2 Primers used in this study.
Table S3 Sequences of Cas9 expression cassettes.
Table S4 Sequences of sgRNA expression cassettes in this study.
Table S5 Sequences of donor DNAs in this study.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Q., Zhao, Q., Liu, Q. et al. CRISPR/Cas9-mediated genome editing in Penicillium oxalicum and Trichoderma reesei using 5S rRNA promoter-driven guide RNAs. Biotechnol Lett 43, 495–502 (2021). https://doi.org/10.1007/s10529-020-03024-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-03024-7