- 1Key Laboratory of Bio-Resources and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, China
- 2School of Landscape and Ecological Engineering, Hebei University of Engineering, Handan, China
- 3Institute of New Energy and Low-Carbon Technology, Sichuan University, Chengdu, China
- 4School of Water Conservancy and Hydroelectric Power, Hebei University of Engineering, Handan, China
Tobacco (Nicotiana tabacum L.) seed lipid is a promising non-edible feedstock for biodiesel production. In order to meet the increasing demand, achieving high seed lipid content is one of the major goals in tobacco seed production. The TT8 gene and its homologs negatively regulate seed lipid accumulation in Arabidopsis and Brassica species. We speculated that manipulating the homolog genes of TT8 in tobacco could enhance the accumulation of seed lipid. In this present study, we found that the TT8 homolog genes in tobacco, NtAn1a and NtAn1b, were highly expressed in developing seed. Targeted mutagenesis of NtAn1 genes was created by the CRISPR-Cas9-based gene editing technology. Due to the defect of proanthocyanidin (PA) biosynthesis, mutant seeds showed the phenotype of a yellow seed coat. Seed lipid accumulation was enhanced by about 18 and 15% in two targeted mutant lines. Protein content was also significantly increased in mutant seeds. In addition, the seed yield-related traits were not affected by the targeted mutagenesis of NtAn1 genes. Thus, the overall lipid productivity of the NtAn1 knockout mutants was dramatically enhanced. The results in this present paper indicated that tobacco NtAn1 genes regulate both PAs and lipid accumulation in the process of seed development and that targeted mutagenesis of NtAn1 genes could generate a yellow-seeded tobacco variety with high lipid and protein content. Furthermore, the present results revealed that the CRISPR-Cas9 system could be employed in tobacco seed de novo domestication for biodiesel feedstock production.
Introduction
Due to the increasing concerns about climate change resulting from excessive consumption of petroleum products, biodiesel has attracted more and more attentions in the recent years. Tobacco (Nicotiana tabacum L.) is an oilseed plant with a high seed lipid content ranging from 36 to 41% of the seed dry weight (Zlatanov et al., 2007; Usta et al., 2011). Recently, tobacco seed lipid had been demonstrated to be a promising feedstock for biodiesel production (Giannelos et al., 2002; Veljkovic et al., 2006; Banković-Ilić et al., 2012; Kumar and Sharma, 2016). Furthermore, due to the rich carbohydrates, wide availability, and low cost, tobacco stalk could also be used for biofuel production (Cai et al., 2016). Recently, a high seed yield tobacco variety, Solaris, had been bred by the Sunchem Holding Company for seed lipid feedstock production (Grisan et al., 2016). Life cycle analysis showed that the impacts created by the production of Solaris tobacco biodiesel were similar to those from other biodiesel plants (Carvalho et al., 2019). Sustainable provision of feedstock is the key to sustainable biofuels (Poltronieri, 2016). In order to meet the huge demand for the feedstock of biodiesel production, achieving high tobacco seed lipid content is one of the main goals in the future.
Sucrose produced by photosynthetic tissues serves as a major carbon source for the synthesis of both seed storage compounds and the generation of other seed components such as mucilage and proanthocyanidins (PAs, also called as condensed tannins) in seed coat. Seed coat development competes for sucrose with reserve component synthesis in embryo and endosperm. Recently, studies had demonstrated that the amount of PAs in the seed coat is negatively correlated with the amount of lipid content in the embryo in Arabidopsis and rapeseed (Wang et al., 2014; Zhai et al., 2019). Thus, strategies of manipulating the PA biosynthesis pathway in seed coat could be used to increase the seed lipid content.
Proanthocyanidin deposit in the inner integument of the seed coat, and the oxidation of PAs in the process of seed maturation results in the formation of brown pigments that confer color to the mature seed (Lepiniec et al., 2006). Previous studies had demonstrated that the biosynthesis of PAs were mainly regulated at the transcription level by transcription factors belonging to the basic helix-loop-helix (bHLH), R2R3-MYB, and WD-repeat protein families (Lepiniec et al., 2006; Xu et al., 2015b). In Arabidopsis, TT2, TT8, and TTG1, which encode R2R3-MYB, bHLH, and WD40 repeat proteins, respectively, form a ternary complex to activate the expression of PA-specific genes during seed coat development (Baudry et al., 2004; Xu et al., 2015b).
Previous studies revealed that TT8 gene played a key regulation role in seed PA biosynthesis. In Arabidopsis, the TT8 gene is required for the expression of DFR and BAN genes in siliques and young seedlings (Nesi et al., 2000). Due to the defect of PAs synthesis, Arabidopsis tt8 mutants showed the transparent test of a phenotype (Nesi et al., 2000). In addition to Arabidopsis, natural mutation of TT8 genes resulted in yellow seed coat trait in allotetraploid Brassica juncea (Padmaja et al., 2014). Most recently, targeted mutation of the TT8 homologs through the CRISPR-Cas9 system in Brassica napus also generated a yellow-seeded phenotype (Zhai et al., 2019). Homologs of TT8 gene were also reported to be involved in PA biosynthesis in diverse plant species, including Medicago truncatula, Lotus corniculatus, and Raphanus sativus (Li et al., 2016; Escaray et al., 2017; Lim et al., 2017).
In addition to its critical role in PA biosynthesis regulation, the Arabidopsis TT8 protein could repress the seed lipid accumulation by inhibiting the expression of transcription factors, including LEC1, LEC2, and FUS3, which play the key roles in embryo development and seed lipid biosynthesis (Chen et al., 2014). Furthermore, TT8 protein could directly repress the expression of genes encoding enzymes involved in fatty acid biosynthesis by binding to the promoter region (Chen et al., 2014). Thus, the mutation of TT8 gene generated seed with a thinner seed coat, a reduced PAs content, and an increased content of lipid in Arabidopsis and Brassica species (Li et al., 2012; Chen et al., 2014; Zhai et al., 2019). We suggested that TT8 gene could be a promising target aimed at enhancing the lipid content for an oilseed plant.
Due to the presence of PAs, tobacco seed shows a dark brown seed color. Thus, we proposed that TT8 homolog genes in tobacco might be an ideal candidate to create a high lipid content seed for biodiesel production. Previous report had identified and characterized two TT8 homolog genes in tobacco genome, NtAn1a and NtAn1b, originated from two ancestors of tobacco, Nicotiana sylvestris and Nicotiana tomentosiformis, respectively (Bai et al., 2011). NtAn1 genes were demonstrated to be involved in flower anthocyanin biosynthesis (Bai et al., 2011); however, if NtAn1 genes regulate the accumulation of PAs and lipid during seed development have not been reported. In this present paper, the expression pattern of NtAn1 genes during seed development was analyzed, and the CRISPR-Cas9 system was applied to generate NtAn1 knockout mutants. We found that the targeted mutagenesis of NtAn1 genes significantly enhanced the accumulation of seed lipid and protein. These results demonstrated that the CRISPR-Cas9-mediated knockout of NtAn1 genes is an efficient approach to improve lipid production in tobacco.
Materials and Methods
Plant Material and Growth Condition
Wild-type (WT) tobacco cultivar (Nicotiana tabacum L. “K326”) was used for gene expression analysis and genetic transformation. Tobacco seeds were surface-sterilized with 75% ethanol for 5 min and washed three times with absolute ethanol. Then, the surface-sterilized seeds were germinated on 1/2 MS medium containing 2% sucrose and 0.7% agar at 25 ± 1°C. The seedlings were transferred to soil and grown in greenhouse at 25 ± 1°C with a photoperiod of 16-h light/8-h dark.
Quantitative Real-Time PCR
Total RNA was isolated using Plant Total RNA Isolation Kit (Foregene, China). PrimeScript™ RT reagent Kit (Takara, Dalian) was used to generate cDNA according to the manufacturer’s instruction. Quantitative real-time PCR (qRT-PCR) was carried out using SYBR Premix Ex Taq™ II (Takara, Dalian) on a CFX96 real-time PCR system (Bio-Rad, United States). The qRT-PCR cycling began with one cycle at 95°C for 2 min, followed by 40 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The house-keeping gene, NtGAPDH (XM_016643257), was used as the reference standard. Three biological and three technical replicates were performed. Primers used for qRT-PCR analysis are listed in Supplementary Table S1.
Vector Construction
To generate knockout mutants using the CRISPR-Cas9 system, the pKSE401 vector (purchased from Addgene: #62202) was used for tobacco genetic transformation. Two guide RNAs (gRNA) targeting the second extron of NtAn1 coding region were designed using CRISPR-P 2.0 (Liu et al., 2017).1 The scaffold containing two gRNAs was amplified by PCR using a pCBC-DT1T2 vector (purchased from Addgene: #50590) as a template. The PCR product was purified using the Universal DNA Purification Kit (TIANGEN, China) and inserted into the pKSE401 vector using a Golden-Gate Assembly method as described previously (Xing et al., 2014). The reconstructed vector was introduced into the DH5α strain of Escherichia coli and confirmed using a Sanger sequencing (Sangon Biotech, China). The verified vector (Named as pKSE401-An1) was introduced into Agrobacterium tumefaciens strain GV3101 for tobacco genetic transformation. Primers used for vector construction are listed in Supplementary Table S2.
Tobacco Transformation and Mutant Selection
Agrobacterium-mediated tobacco leaf disc transformation experiment was performed as previously described (Horsch, 1985). T0 generation transgenic lines were selected on mass spectrometry (MS) medium supplemented with 50 mg/l kanamycin. Genomic DNA was extracted from the kanamycin-resistant T0 transgenic lines using a Super Plant Genomic DNA Kit (TIANGEN, China). To select mutant lines, the flanking region of the gRNAs targeting sites was amplified using sequence-specific primers by PCR (primers are listed in Supplementary Table S2). The PCR products were purified and sequenced immediately by the Sanger method (Sangon Biotech, China). The T0 mutant lines were self-pollinated to generate T1 seeds. The T1 plants were analyzed again to confirm the mutation. In addition, the presence of the CRISPR-Cas9 construct in T1 mutant plants was examined by PCR using vector specific primers (primer sequences are listed in Supplementary Table S2). The homozygous T1 mutant plants without the CRISPR-Cas9 construct were used for further analysis.
DMACA Staining
To visualize the accumulation of PAs in seed coat, dry mature tobacco seeds from WT and mutant plants were stained in a freshly prepared dimethylaminocinnamaldehyde (DMACA, Sigma, United States) reagent [2% (w/v) DMACA dissolved in 6 N HCl/95% ethanol mixture (1:1, v/v)] for 30 min and then washed several times with 70% ethanol (v/v) as described previously (Hong et al., 2017). The stained seeds were photographed using a Leica stereomicroscope (Leica, Germany).
PAs Quantification
Quantification of PAs was performed according to the previous report (Pang et al., 2008). Briefly, to extract soluble PAs from tobacco seeds, 200 mg of dry seeds were ground in liquid nitrogen and extracted with 1 ml extraction solution (70% acetone/0.5% acetic acid) by vortexing for 10 s. After sonication at room temperature for 1 h, the mixture was centrifuged at 2,500 g for 10 min, and the residue was re-extracted twice. The pooled supernatants were extracted twice with hexane. To quantify the soluble PAs level, 50 μl of the supernatant sample was mixed with 200 μl of DMACA reagent (0.1% DMACA, 90% ethanol, 10% HCl) in 96-well plates, and the absorption was measured at 640 nm. Soluble PA levels were calculated using a standard curve prepared using procyanidin B1 (Sigma, United States).
The residue from soluble PAs extraction was air dried and used for quantitative analysis of insoluble PAs. Five-hundred microliter butanol-HCl reagent (95% butanol:5% concentrated HCl) was added to the residue, and the mixture was sonicated at room temperature for 1 h, followed by centrifugation at 2,500 g for 10 min. The absorption of the supernatant was measured at 550 nm, then samples were boiled for 1 h and cooled to room temperature, and the absorbance at 550 nm was recorded again, with the first value being subtracted from the second. Absorbance values were converted into PA equivalents using a standard curve of procyanidin B1 (Sigma, United States).
Seed Lipid Assay
To determine the seed lipid content and fatty acid composition, fatty acid methyl esters (FAMEs) were prepared as previously described (Li et al., 2006). Twenty mature tobacco seeds were added into the methyl esterification solution [1 ml of 5% sulfuric acid in methanol (v/v), 25 μl 0.2% butylated hydroxyl toluene solution, and 300 μl toluene. Twenty microgram triheptadecanoin was added as internal standard]. The mixture was heated at 90°C for 2 h. Then, 1.5 ml of 0.9% NaCl and 1 ml hexane were added after the mixture had cooled down to room temperature. The FAMEs were separated by collecting the organic phase. The FAMEs were quantitatively analyzed by gas chromatography mass spectrometry (GC–MS; DAOJING, Japan). The GC conditions were as follows: 1 μl injection volume, split injection (1:20), injector temperature 220°C, oven temperature program: 150°C for 1 min, then increased to 200°C at 10°C min−1, holding at 200°C for 1 min, and then increased to 210°C at 5°C min−1 and held for 1 min. The seed lipid content was quantified according to the peak area of the internal standard.
Protein Assay
To evaluate the seed protein content, the total protein was extracted as previously described with some modification (Heath et al., 1986). Briefly, 10 mature tobacco seeds were grounded in protein extraction solution [63 mM Tris buffer, pH 7.8, 0.5 M NaCl, and 0.07% (v/v) β-mercaptoethanol]. The homogenate samples were centrifuged at 12,000 rpm for 10 min. Twenty microliter of the supernatant was used for protein quantification using the Pierce BCA Protein Assay Kit (Thermo, United States) according to the manufacturer’s protocol. The experiment was repeated three times.
Yeast One-Hybrid Assay
Yeast one-hybrid assay was performed to investigate if An1b protein could bind to the promoter of the anthocyanidin reductase (ANR) gene. The promoter sequence of ANR gene (574 bp upstream of the start codon) was amplified by PCR using primers pANR-F and pANR-R (primer sequences were listed in Supplementary Table S2) and inserted into the yeast expression vector pHIS2 using the ClonExpress II One Step Cloning Kit (Vazyme, China) to generate pHIS2-ANR. The An1b protein coding region was amplified by PCR using An1b-F and An1b-R (primer sequences are listed in Supplementary Table S2) and cloned into the pGADT7 vector using the ClonExpress II One Step Cloning Kit (Vazyme, China) to generate pGADT7-An1b. The coding region of An2 protein was also amplified using An2-F and An2-R (primer sequences are listed in Supplementary Table S2) and firstly inserted into pDR195 vector using the ClonExpress II One Step Cloning Kit (Vazyme, China). Then, the An2 expression box in pDR195 vector was amplified using An2-box-F and An2-box-R (primer sequences are listed in Supplementary Table S2), and the fragment was inserted into pHIS2-ANR using the ClonExpress II One Step Cloning Kit (Vazyme, China) to obtain pHIS2-ANR + An2. The yeast strain Y187 was co-transformed with pGADT7- and pHIS2- based vectors by the polyethylene glycol-mediated method. The DNA-protein interactions were evaluated according to the growth status of yeast cells cultured on the SD/−Leu/−Trp/-His selective medium with 120mM 3-AT for 3 days at 30°C.
Yield-Related Traits Assay
For seed size measurement, mature seeds were photographed using a Leica stereomicroscope (Leica, Germany). The seed length and width were measured with the Image J software. For average seed weight analysis, 50 seeds were randomly collected and carefully weighed using an electronic balance (METTLER TOLEDO, United States). Average seed weight was calculated by dividing the total seed weight by the seed number. Fruit number per plant was checked from 20 individual plants at the mature stage. Fruits used for the seed number analysis were obtained from the first five basal fruits of the main inflorescence. The total seed weight from a single fruit was weighed using an electronic balance, and the seed number per fruit was calculated by dividing the total seed weight by the average seed weight.
Results
NtAn1 Genes Were Highly Expressed in Developing Seed
Previous study reported that Arabidopsis TT8 gene has two homologs in tobacco genome, NtAn1a and NtAn1b (Bai et al., 2011). Tobacco is a natural allotetraploid plant; the sequence analysis revealed that NtAn1a originated from N. sylvestris, whereas NtAn1b derived from N. tomentosiformis. The qPCR analysis showed that NtAn1a and NtAn1b expressed at developing flowers with the highest expression level in corolla limb, which was consistent with the function of the flower flavonoid biosynthesis regulation (Bai et al., 2011). However, from the previously reported results, we noticed that the transcript level of both NtAn1a and NtAn1b was relatively high in developing ovary, which indicated that NtAn1 genes might play an important role in seed development (Bai et al., 2011).
To further confirm and characterize the expression pattern of NtAn1a and NtAn1b, their expression was assessed in different organs and various stages of seed development: 7, 14, 21, and 28 days after flowering (DAF). qRT-PCR results showed that NtAn1a and NtAn1b had a similar expression pattern, with the highest expression in developing seeds at 7 DAF and decreased at later stages (Figure 1). The expression of both NtAn1 genes exhibited a high expression level in flower, which was consistent with the previous results. However, low transcript levels were detected in root, leaf, and stem (Figure 1). These results suggested that NtAn1a and NtAn1b might regulate PAs and lipid accumulation during seed development in a way like in Arabidopsis and rapeseed.
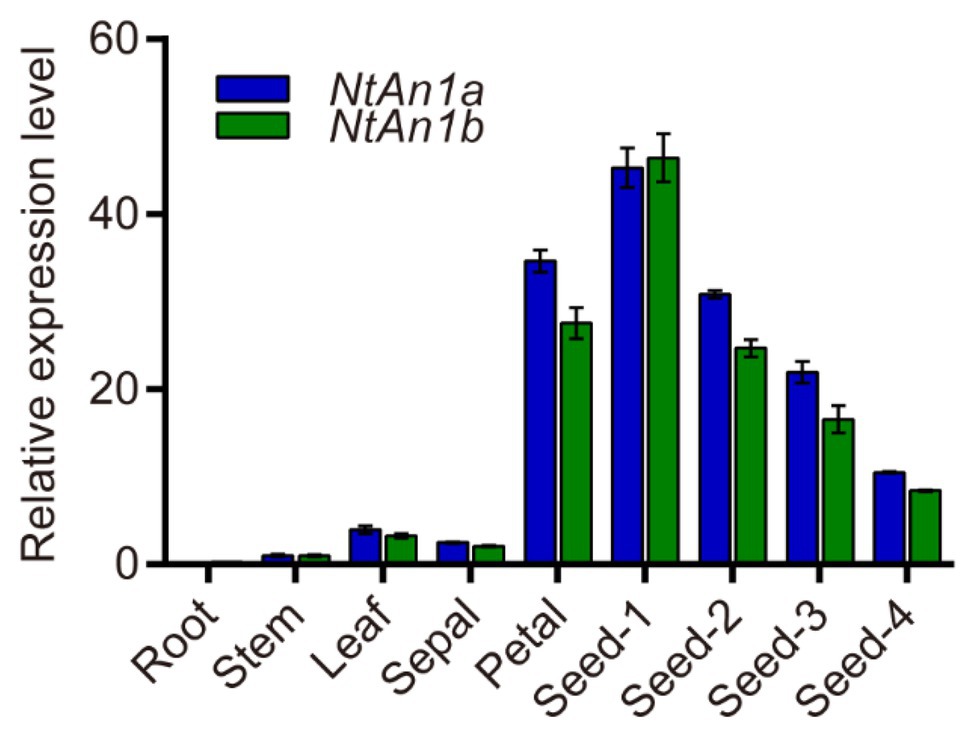
Figure 1. Expression profiles of tobacco NtAn1a and NtAn1b genes. Seed-1, developing seed at 7 days after flowering (DAF); Seed-2, developing seed at 15 DAF; Seed-3, developing seed at 21 DAF; Seed-4, developing seed at 28 DAF. Data are mean ± SD (n = 3). NtGAPDH gene was amplified as an internal control.
Targeted Mutagenesis of NtAn1 Using the CRISPR-Cas9 System
NtAn1a and NtAn1b showed a high sequence identity of 92.95 and 90.36% at the nucleotide and protein levels, respectively (Supplementary Figures S1, S2). Furthermore, these two genes had similar expression patterns (Figure 1). These results suggested that these two genes may have similar and redundant functions. Thus, two gRNAs recognizing both NtAn1 genes were designed to effectively knockout them and both of the gRNAs targeting the second extron of the coding sequence (Figure 2A). The CRISPR-Cas9 construct containing these two gRNAs, which were driven by the Arabidopsis U6-26 and U6-29 promoter, respectively (Figure 2B), was produced based on the CRISPR-Cas9 multiplex genome-editing vector (Xing et al., 2014). The resulting construct was transformed into WT tobacco plant using the Agrobacterium-mediated leaf disc transformation method. Through kanamycin selection, 12 kanamycin resistance T0 transgenic plants were generated. The targeting region of both NtAn1a and NtAn1b were amplified by a pair of primers at the same time. Two homozygous mutant lines (an1-1 and an1-2) were identified from the T0 transgenic plants by Sanger sequencing analysis of the gRNAs targeting region. The an1-1 mutant had one base insertion at both gRNA targeting sites of the NtAn1a and NtAn1b genes, while the an1-2 mutant line had a 105 base fragment deletion between the two gRNAs targeting sites (Figures 2C,D). T-DNA free mutant plants were selected from the T1 progeny generated by the self-pollinated of two independent T0 homozygous mutant lines. Twenty T-DNA free T1 generation plants from each mutant line were randomly selected for further analysis.
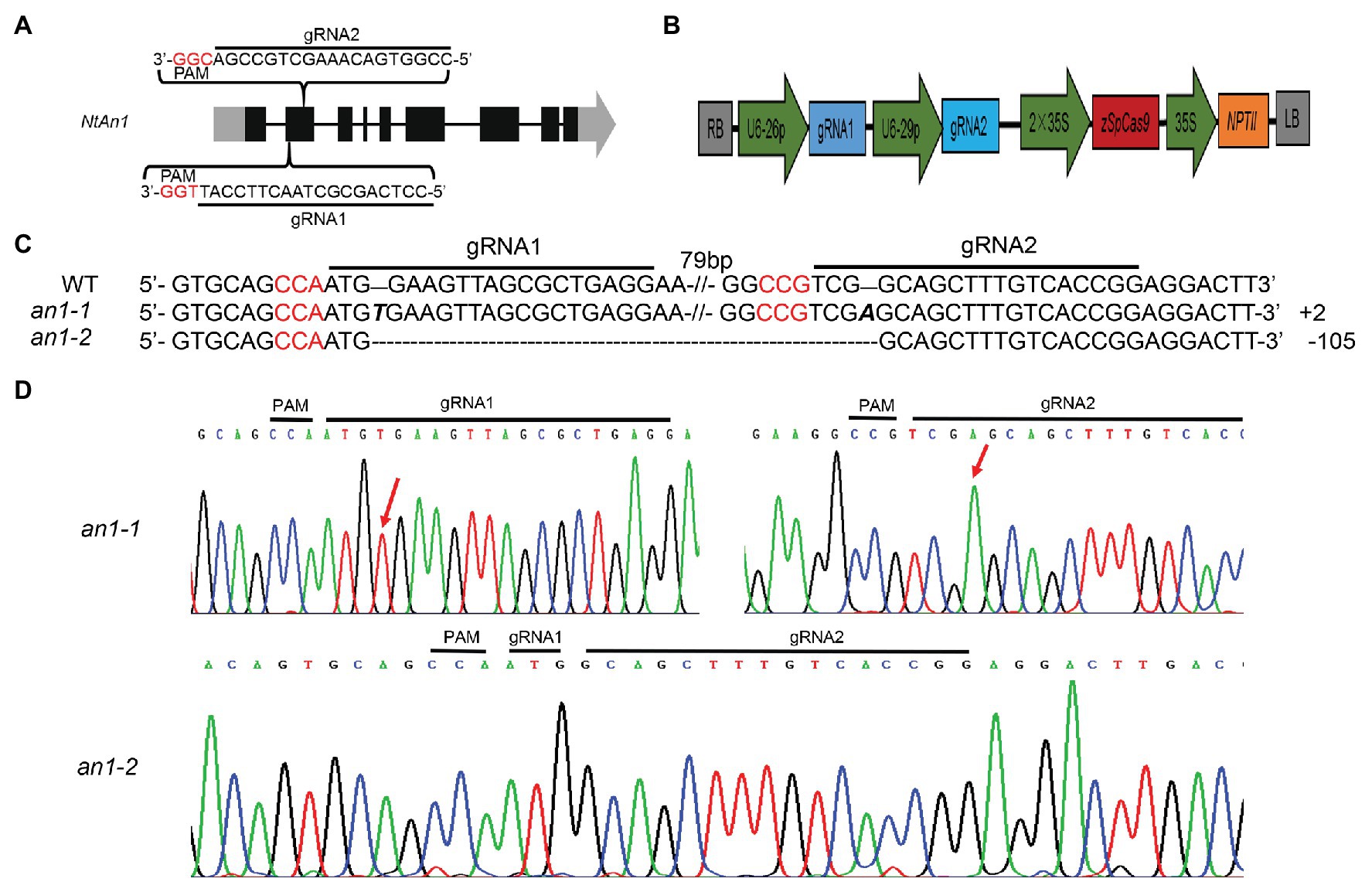
Figure 2. CRISPR-Cas9-mediated targeted mutation of NtAn1. (A) Targeting sites and gRNA sequences used for NtAn1 genes editing. The three base protospacer adjacent motif (PAM) was in red. (B) The CRISPR-Cas9 vector structure used for targeted mutation of NtAn1. (C) Mutation type of the an1-1 and an1-2 lines determined by the Sanger method. (D) Sanger sequencing peaks of the an1-1 and an1-2 mutant lines. The red arrows indicate the location of mutations.
Mutation of NtAn1 Genes Resulted in Yellow Seed Coat
The formation of seed color in most plant species is due to the deposition of PAs within the endothelial layer of the inner integument of the seed coat (Lepiniec et al., 2006). Previous studies demonstrated that TT8 played a key role in regulating PAs accumulation in various plants (Li et al., 2016; Escaray et al., 2017). In this paper, mutation of the NtAn1, the TT8 homolog genes in tobacco, generated a yellow-seeded phenotype (Figure 3A). This indicated that the targeted mutation of the NtAn1 genes might disrupt the accumulation of PAs in tobacco seed coat. In order to check the PAs deposition visibly, the mutant tobacco seeds were dyed by DMACA reagent. The results further confirmed the defects of PAs accumulation in seed coat (Figure 3B). The soluble and insoluble PAs contents were calculated quantitatively by spectrophotometric method. The results showed that the PAs were mainly stored in the insoluble form in tobacco seed coat, and the targeted mutation of NtAn1 genes led to the significant decreases in both soluble and insoluble PAs content compared with those in WT tobacco seed coat (Figure 3C).
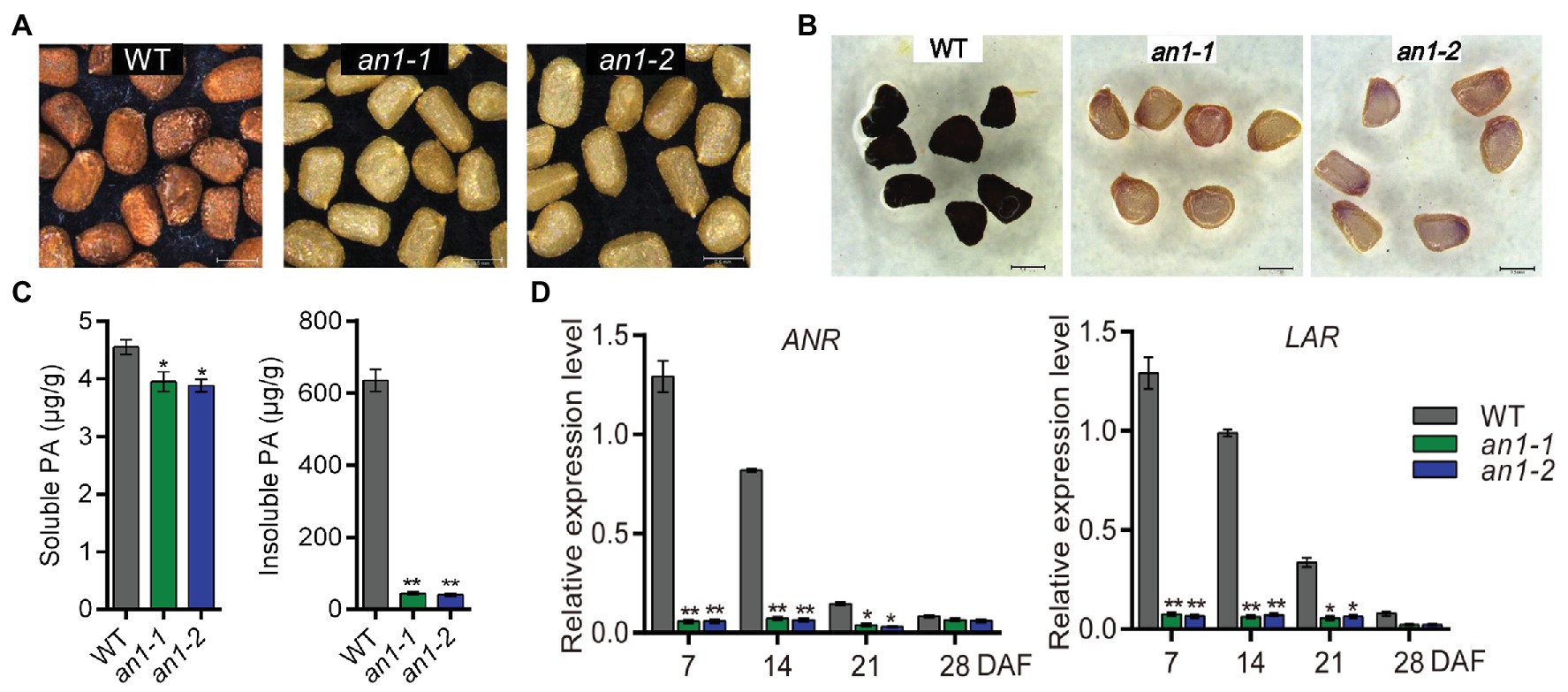
Figure 3. Seed phenotypes of tobacco an1 mutants. (A) Tobacco seed coat color phenotypes of WT and an1 mutant lines. (B) DMACA staining of mature tobacco seeds from WT and an1 mutant lines. (C) Soluble and insoluble PA contents in mature seeds of WT control and an1 mutant lines. Three biological replicates were analyzed. Data are mean ± SD (n = 3). (D) Expression of PA biosynthetic genes regulated by NtAn1 in tobacco. ANR: anthocyanidin reductase and LAR: leucoanthocyanidin reductase. RNA samples were extracted from the developing seeds at 7, 14, 21, and 28 DAF. Data are mean ± SD of three biological replicates. Asterisks indicate significant difference compared to control samples (*p < 0.05; **p < 0.01). NtGAPDH gene was used as an internal reference.
ANR and leucoanthocyanidin reductase (LAR) are two key enzymes participated in PA biosynthesis. ANR converts anthocyanidins to the epicatechin (Xie et al., 2003), whereas LAR could reduce leucocyanidin to catechin (Tanner et al., 2003). Catechin and epicatechin are considered to be the main building blocks for PAs biosynthesis. The encoding genes of ANR and LAR were directly regulated by TT8 gene in Arabidopsis (Baudry et al., 2004). The expression patterns of the homolog genes encoding these two enzymes during seed development were analyzed by qRT-PCR. Our results showed that both of these two genes had a similar expression profile as NtAn1, and the expression of them were significantly decreased in both an1-1 and an1-2 mutant seed (Figure 3D). Taken together, these findings indicate that tobacco NtAn1 regulated the accumulation of PAs in a similar way like in Arabidopsis, and the mutation of the tobacco NtAn1 genes could hinder the PA deposition in the seed coat, which was consistent with the phenotypes observed in tt8 mutant seed in Arabidopsis and other Brassica species (Li et al., 2012; Chen et al., 2014; Padmaja et al., 2014; Zhai et al., 2019).
In order to confirm if the NtAn1 protein could directly bind to the promoter region of the genes involved in PA biosynthesis, yeast one-hybrid assay was performed. The promoter region of ANR gene was inserted into the pHIS2 to generate ppHIS2-LAR as a reporter vector (Figure 4A). Due to the high protein sequence identity of NtAn1a and NtAn1b, the NtAn1b protein was chosen to test the DNA binding ability (Figure 4A). Our result showed that the NtAn1b protein could not bind to the ANR promoter region in yeast cells and activate the HIS reporter gene alone (Figure 4B). Previous study demonstrated that NtAn1 proteins could interact with tobacco An2, a R2R3-MYB family protein (Bai et al., 2011). In addition, the bHLH protein usually forms a MYB-bHLH-WD40 (MBW) ternary complex with WD40 and R2R3-MYB proteins to regulate the expression of downstream PA biosynthesis genes (Baudry et al., 2004; Schaart et al., 2013; Xu et al., 2015b). We speculated that the regulation activity of NtAn1 protein might be dependent on An2. To verify this speculation, the An2 protein was expressed using the pHIS2 vector by introducing an expression box from pDR195 to generate pHIS2-ANR + An2 (Figure 4A). When pGADT7-An1b and pHIS2-ANR + An2 were co-transformed into yeast cells, the HIS reporter gene could be activated (Figure 4B). These results indicated that NtAn1b protein functions together with An2 protein in PA pathway gene regulation. However, the WD40 protein has not been identified so for.
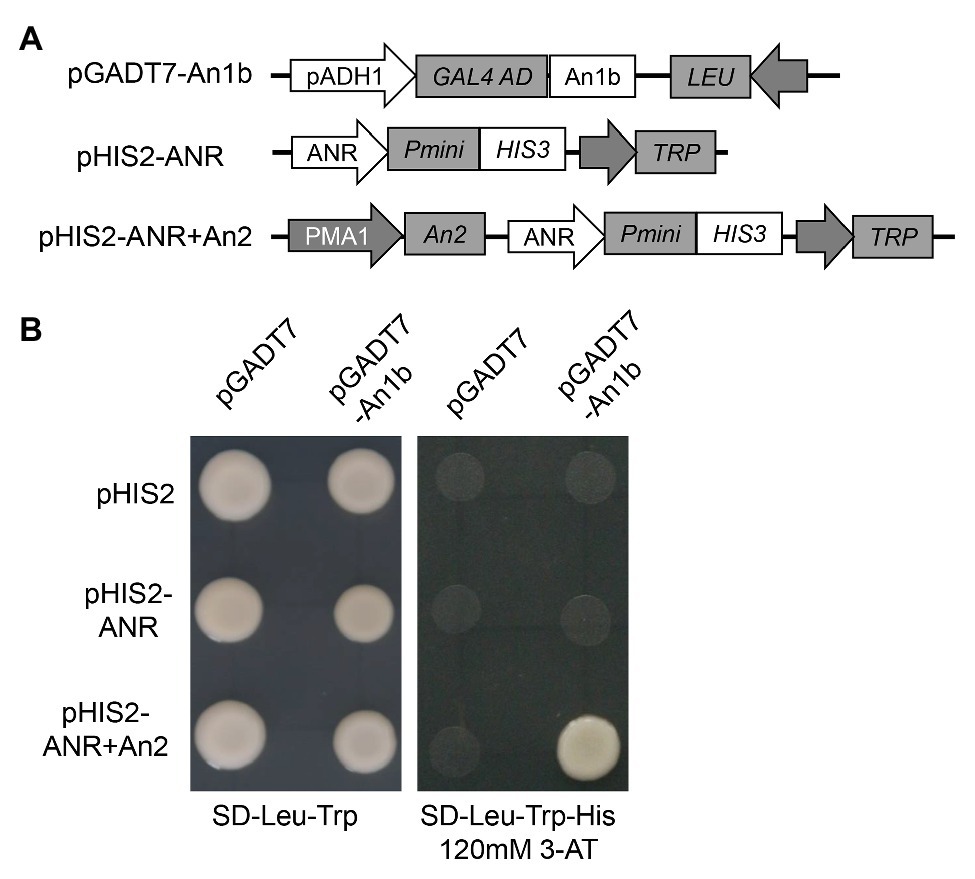
Figure 4. NtAn1 Yeast one-hybrid assay. (A) Schematic structure of the vectors used for yeast one-hybrid assay. (B) Growth performance on transformed yeast cells on SD/−Leu−/Trp and SD/−Leu−/Trp/-His medium containing 120mM 3-AT.
Targeted Mutagenesis of NtAn1 Generated White Flower
Previous study had demonstrated that the anthocyanin accumulation in transgenic tobacco flowers could be significantly elevated by the overexpression of NtAn1a or NtAn1b gene (Bai et al., 2011). The early biosynthesis genes (EBGs), such as CHS, CHI, and F3H, and the late biosynthesis genes (LBGs) in the anthocyanin pathway, including DFR and ANS, were dramatically induced by the overexpression of NtAn1 genes in tobacco (Bai et al., 2011). In this present paper, we found that the targeted mutagenesis of NtAn1 genes generated white flower phenotype, which resulted from the defects in anthocyanin accumulation in the flower (Figure 5A). Previous study demonstrated that TT8 gene regulated the biosynthesis of anthocyanin by manipulating the expression of the LBGs in anthocyanin pathway (Zhai et al., 2019). The expression level of the downstream genes at different flower development stages were analyzed by qRT-PCR. Our results revealed that the examined anthocyanin biosynthesis genes expressed at all three developmental stages with the expression level peaking at the late stage in WT plant flower (Figure 5B). The expression patterns of these genes were consistent well with those of NtAn1 genes, which indicated the regulation relation between them (Bai et al., 2011). By contrast, the expression of all examined anthocyanin biosynthesis genes at different developing stages was significantly repressed in the an1-1 mutant line (Figure 5B). Taken together, our results demonstrated again that NtAn1 genes played an essential role in the biosynthesis of anthocyanin in tobacco flower.
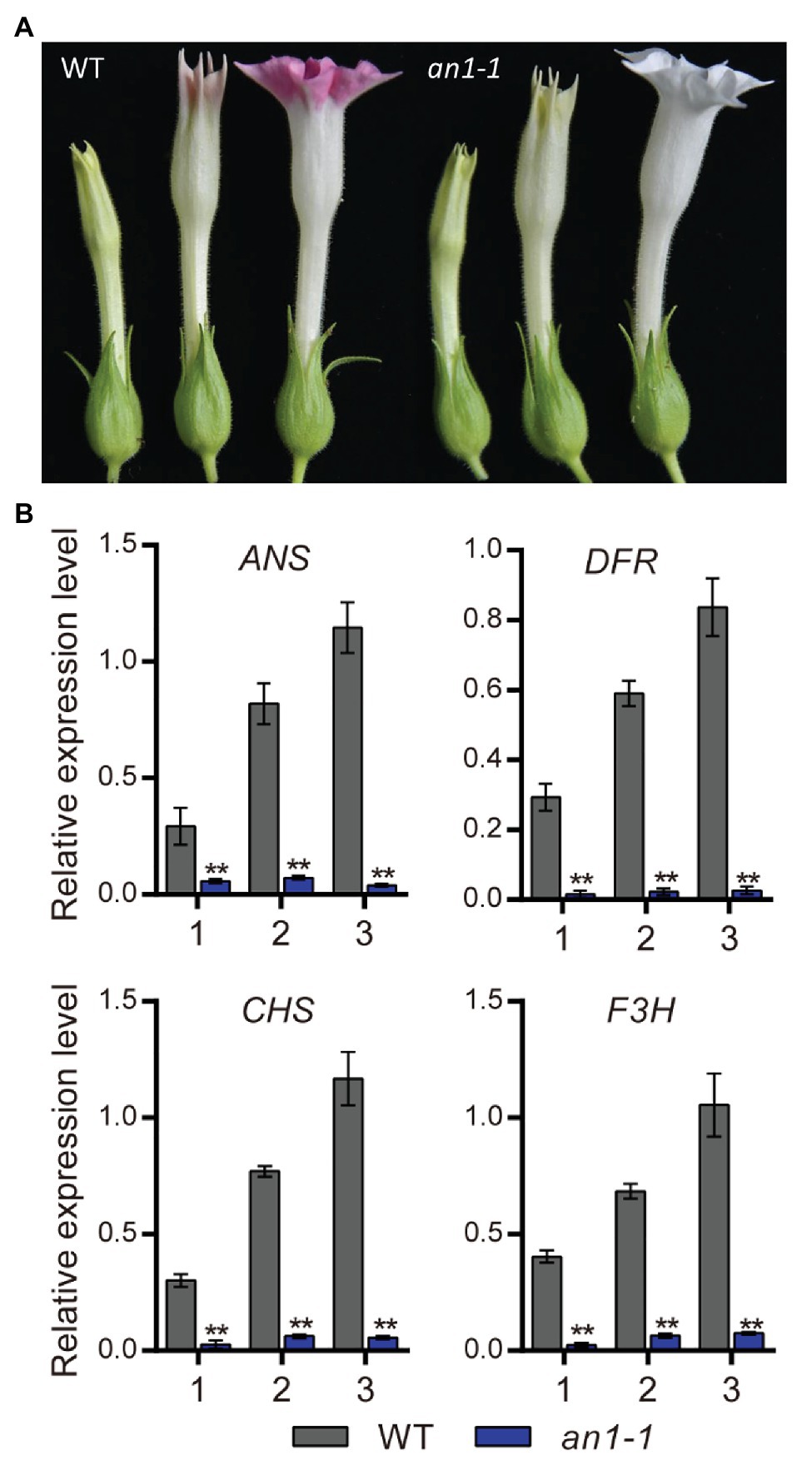
Figure 5. Flower phenotype of tobacco an1-1 mutant. (A) Flower at different development stages from WT and an1-1 mutant line. (B) Expression analysis of anthocyanin biosynthetic genes regulated by NtAn1 in tobacco. CHS, chalcone synthase; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase. Data are mean ± SD (n = 3). Asterisks indicate significant difference compared to control samples (*p < 0.05; **p < 0.01). NtGAPDH gene was used as an internal reference.
Targeted Mutagenesis of NtAn1 Increased Seed Lipid and Protein Content
Both natural and targeted mutation of TT8 genes in Arabidopsis and Brassica species would result in a significant increase in seed lipid content. To characterize the effect of the NtAn1 gene targeted mutation on tobacco seed lipid accumulation, the seed lipid content was analyzed by the GC-MS method. The results showed that WT tobacco seed lipid content was about 38.77 μg per seed, and the lipid content was approximately 45.91 μg per seed in an1-1 and 44.97 μg per seed in an1-2 mutant line, which increased significantly by 18.42 and 15.99% relative to the WT seeds, respectively (Figure 6A). These results indicated that the TT8 gene and its homologs regulated seed lipid accumulation in a conserved way among different plant species. Thus, the TT8 homologs from other oilseed plants could be used as a target to enhance the seed lipid content.
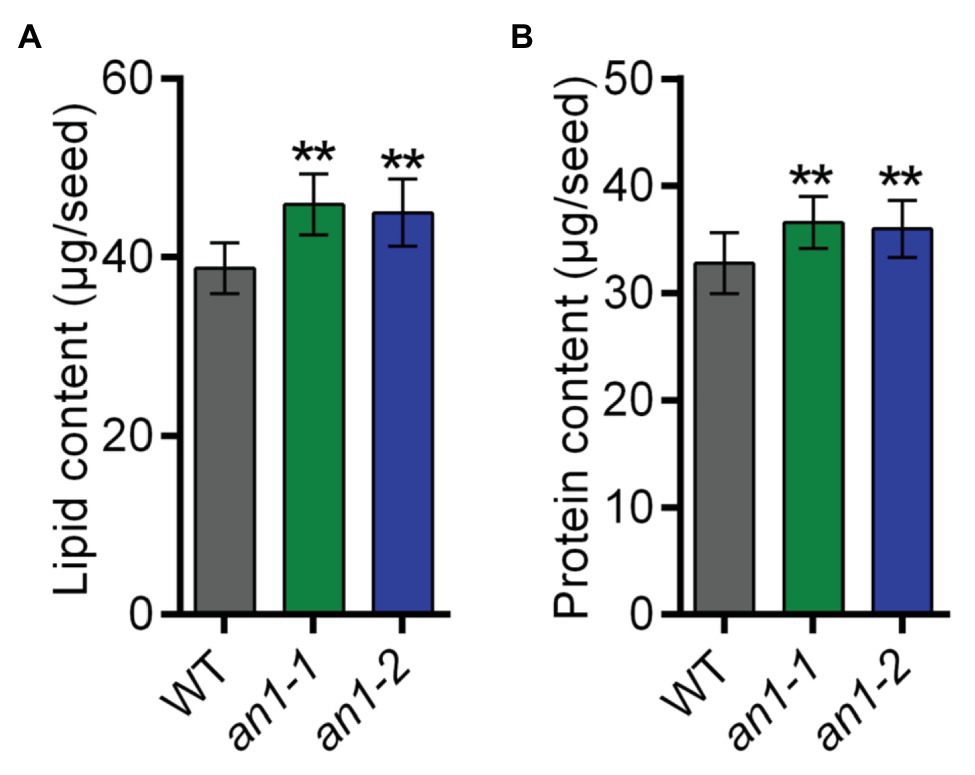
Figure 6. Lipid and protein content of the WT and an1 mutant seeds. (A) Lipid content per seed. Twenty seeds were analyzed for each repeat and per seed lipid content was calculated by dividing the seed number. Data are mean ± SD (n = 3). (B) Protein content per seed. Ten seeds were analyzed for each repeat and per seed protein content was calculated by dividing the seed number. Data are mean ± SD (n = 3). Asterisks indicate significant difference compared to control samples (**p < 0.01).
In most oilseed crops, the seed lipid content is negatively correlated with the protein content. Surprisingly, the BnTT8 mutant seeds showed simultaneous increases in both lipid and protein contents (Zhai et al., 2019), which are different from the lower protein content in Arabidopsis tt8 mutant seed (Chen et al., 2014). These results indicated that the seed protein accumulation was regulated by different mechanism in Arabidopsis and B. napus. To determine the effect of NtAn1 genes on tobacco seed protein accumulation, the seed protein contents in WT and an1 mutant lines were examined using the Pierce BCA Protein Assay Kit. The results showed that WT tobacco seed protein content was about 32.79 μg per seed, and the protein content was elevated to 36.56 μg per seed in an1-1 and to 35.97 μg per seed in an1-2 mutant line, increased significantly by 11.50 and 9.70% relative to the WT seeds, respectively (Figure 6B). These results were consistent with those in B. napus (Zhai et al., 2019). The seed meals of oilseeds are usually used as animal feed. Compared with the black-seeded rapeseed, the increased protein level makes the seed meal generated from yellow-seeded rapeseed more valuable for animal feed production (Bell, 1993; Jiang et al., 2015). The increased protein content of the an1 mutant seeds could make the yellow-seeded tobacco seed meal more valuable for animal feed.
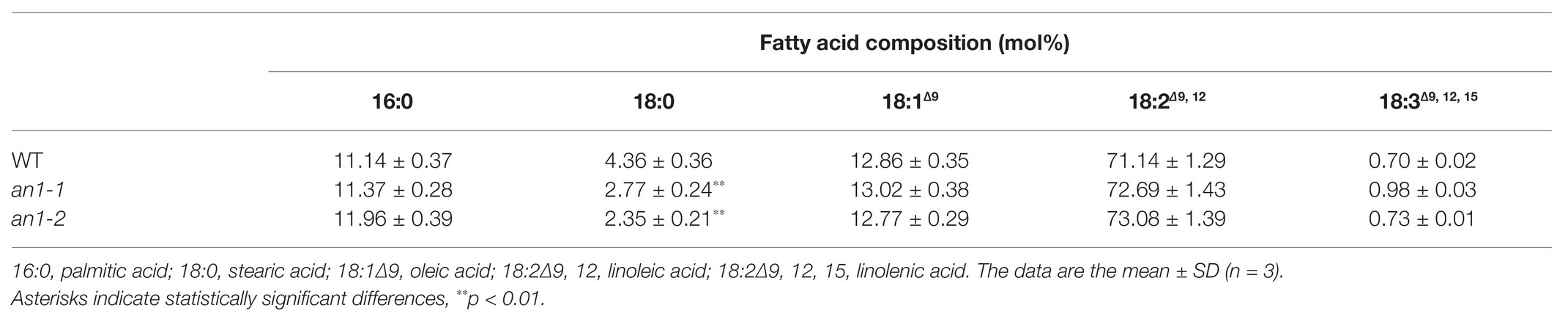
The property of biodiesel is partially determined by the fatty acid chain length, the position, and the number of the double bonds (Durrett et al., 2008). In Arabidopsis and B. napus, the mutation of TT8 gene resulted in an alteration in the seed fatty acid profile, including increases in palmitic acid, linoleic acid, and linolenic acid and decreases in stearic acid and oleic acid compared with the WT seeds (Chen et al., 2014; Zhai et al., 2019). Fatty acid composition of tobacco seed lipid shows the main presence of palmitic acid, stearic acid, oleic acid, and linoleic acid (Giannelos et al., 2002). Possibly due the differences in the fatty acid composition, targeted mutation of NtAn1 genes just resulted in a significant decrease in stearic acid, and the other four main fatty acid components were not changed significantly compared with the WT tobacco seed (Table 1).
Expression of Genes Involved in Seed Development and Lipid Biosynthesis Were Altered by NtAn1 Targeted Mutation
In Arabidopsis, TT8 protein could repress the lipid biosynthesis pathway by directly binding to the promoter region of the critical transcriptional factors, such as LEAFY COTYLEDON1 (LEC1), LEC2, and FUSCA3 (FUS3), that are important for seed development. In addition, TT8 gene could indirectly inhibit the expression of a number of important genes, including KASII, MOD1, FAB2, FatA, FAE1, FAD2, and FAD3, in the fatty acid biosynthesis pathway. Thus, the Arabidopsis tt8 mutant showed increased seed lipid content (Chen et al., 2014). In B. napus L., the expression levels of several genes involved in fatty acid biosynthesis during seed development were increased in BnTT8 mutant plants generated by the CRISPR-Cas9 system (Zhai et al., 2019). In this present paper, the targeted mutagenesis of NtAn1 genes led to the increases of both lipid and protein content, so we suggested that NtAn1 genes might regulate the seed development and storage component accumulation in a way similar to that in B. napus. To test this hypothesis, the expression of the transcription factors regulating seed development, including LEC1, LEC2, FUS3, and the enzymes that are important in the fatty acid biosynthesis pathway, such as KASI, PI-PKβ1, and BCCP2, were examined by qRT-PCR in the process of seed development. Our results showed that the expression of all examined genes was upregulated in the an1-1 mutant at one or two developmental stages (Figure 7). For example, the LEC1 and FUS3 genes were significantly increased at 14 and 21 DAF seeds, while the KASI and PI-PKβ1 genes were upregulated at 14 DAF stage (Figure 7). The elevated expression levels of genes involved in seed development and fatty acid biosynthesis could explain the enhanced lipid content in mutant lines.
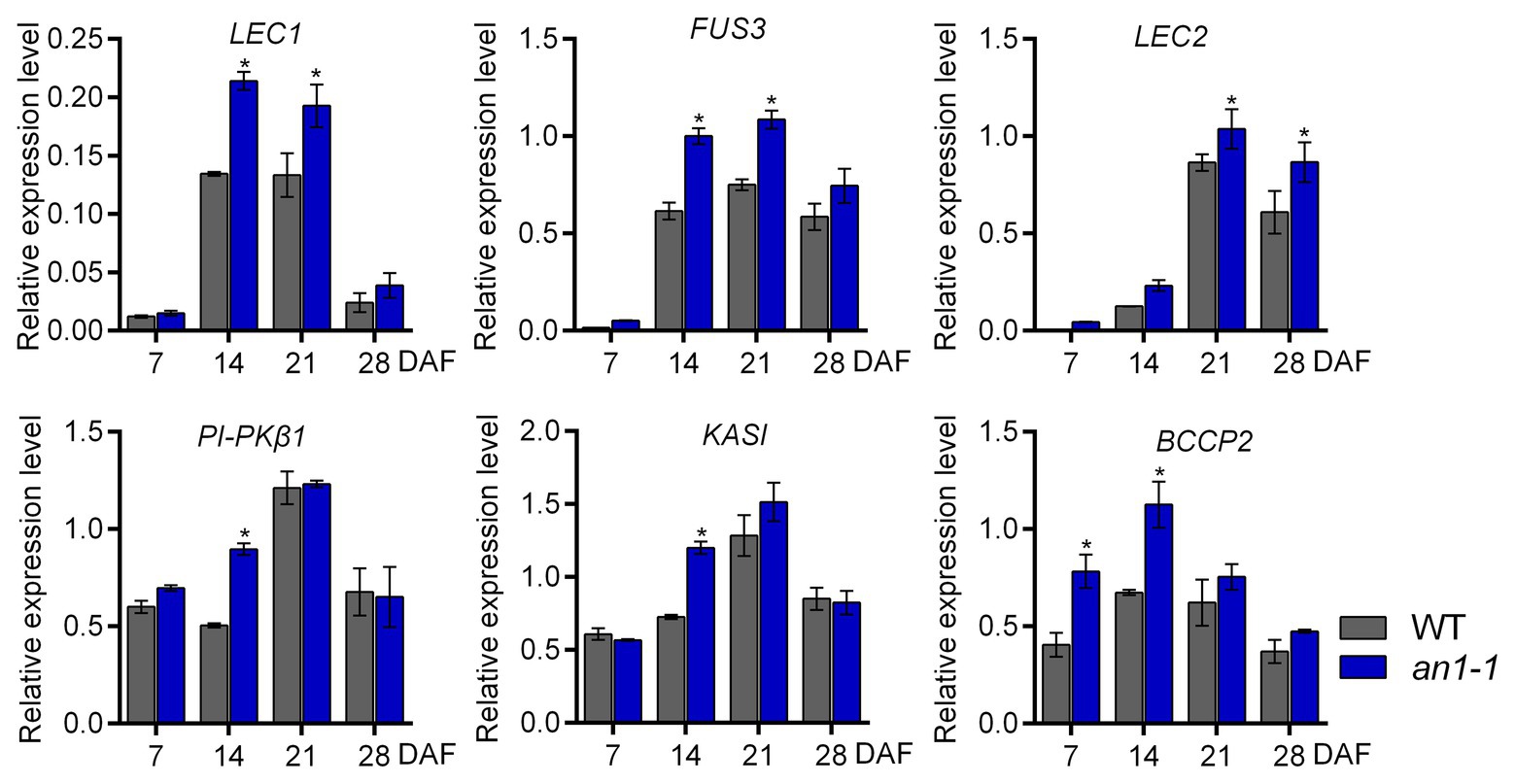
Figure 7. Expression analysis of genes involved in seed development and fatty acid biosynthesis. Asterisks indicate significant difference compared to control samples (*p < 0.05).
Seed Yield Related Traits Were Not Affected by Mutation of NtAn1
Besides the seed lipid content, seed yield is another important factor affecting the lipid yield. The seed yield-related traits of the an1-1 and an1-2 mutant lines were also evaluated. The results showed that the yield-related traits, including seed size (Figures 8A,B), seed weight (Figure 8C), fruit number per plant (Figure 8D), and seed number per fruit (Figure 8E), were not affected by targeted mutation of NtAn1 genes. Thus, the targeted mutation of NtAn1 genes could generate a useful tobacco variety with a high seed lipid yield and improved nutritional quality.
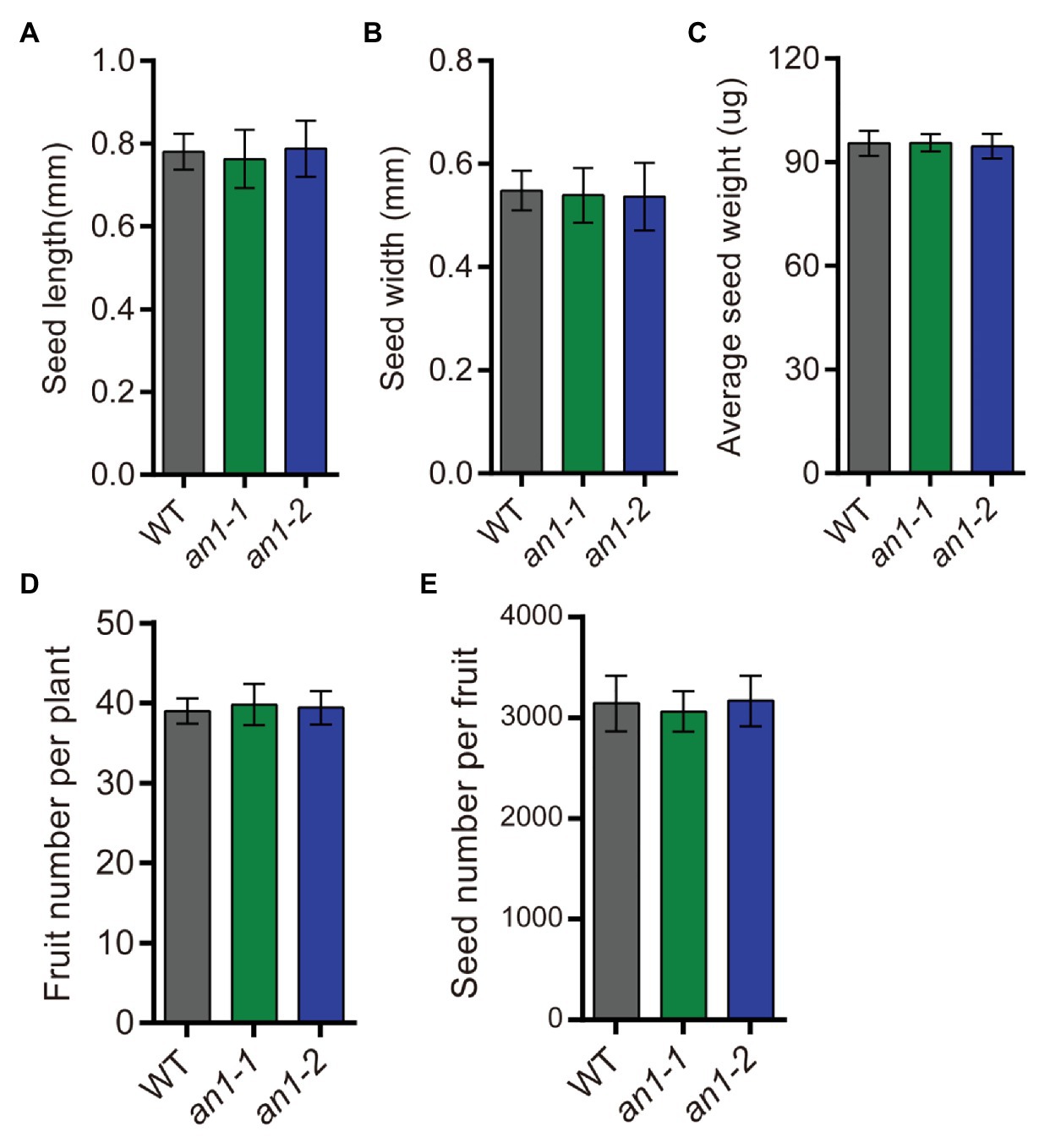
Figure 8. Seed yield related traits of an1 mutant tobacco lines. (A) Average seed length. Data are mean ± SD (n = 30). (B) Average seed width. Data are mean ± SD (n = 30). (C) Average seed weight. Values are mean ± SD of five individual measurements of 50 seeds/replicate. The seeds used to measure seed size and seed weight were selected randomly after collecting all the mature seeds. (D) Fruit number per plant. Fruit number was calculated at the plant mature stage. Data are mean ± SD (n = 20). (E) Seed number per fruit. Data are mean ± SD (n = 10). Seed number per fruit was calculated from the basal five fruits at the mature stage.
Discussion
TT8 Homolog Genes Function in a Conserved Way to Regulate Seed Coat PAs and Seed Lipid Accumulation
Compared with the oilseeds with a black seed coat, the oilseeds with a yellow-seeded phenotype showed a thinner seed coat, a reduced PAs content, and an increased content of lipid, thus a yellow-seeded phenotype is a preferred trait for numerous oilseed plant genetic breeding (Rahman and Mcvetty, 2011). Due to the accumulation of PAs in the seed coat, tobacco seed showed a black color. Previous studies had demonstrated that the TT8 protein played a key regulation role in PA biosynthesis in seed coat (Xu et al., 2014). Two TT8 homologs, NtAn1a and NtAn1b, had been identified in tobacco genome (Bai et al., 2011). However, the functions of seed color formation and seed lipid accumulation of these two genes had not yet been characterized. In this paper, we found that NtAn1 genes were highly expressed in developing seeds (Figure 1), which indicated that they might play a role in seed development. A yellow-seeded phenotype was generated by CRISPR-Cas9-mediated targeted mutagenesis of tobacco NtAn1 genes (Figure 3A). DMACA staining of seed coat and extraction analysis further confirmed that the mutation of NtAn1 genes blocked the specific PA deposition in the seed coat (Figures 3C,D). Most importantly, seed lipid and protein contents were both dramatically increased by targeted mutagenesis of NtAn1 genes (Figure 6). In Arabidopsis, TT8 induces the expression of LBGs of PAs through directly binding to the regulatory region (Xu et al., 2014). Natural mutation of BrTT8 by a large insertion resulted in yellow seed coat in Brassica rapa, and the LBGs were significantly downregulated by BrTT8 mutation (Li et al., 2012). In this paper, the LBGs, including ANR and LAR, were significantly decreased in both an1-1 and an1-2 mutant lines (Figure 3D). In Arabidopsis, TT8 protein could also directly bind to the promoter region of transcription factors that are important for seed development and lipid biosynthesis and repress the expression of them (Chen et al., 2014). In this paper, the expression of the downstream transcription factors and fatty acid biosynthesis genes were significantly upregulated during seed development in the an1-1 mutant line (Figure 7). Most recently, similar phenomena were observed in CRISPR-Cas9-mediated TT8 gene mutation in B. napus (Zhai et al., 2019). Previous reports and results in this present paper indicated that TT8 homolog gene functions in a conserved way in seed coat PAs and seed lipid biosynthesis regulation. Based on the results in this present paper, a model for NtAn1 protein-mediated regulation of PAs and lipid accumulation was constructed (Figure 9). In WT tobacco seed, the NtAn1 protein could inhibit the expression of transcription factor (including LEC1 and FUS3) and genes encoding fatty acid biosynthesis enzyme (including KASI and BCCP2). Due to the inhibition effect of NtAn1, WT seeds showed a lipid content of about 38 μg per seed (Figure 6A), and a protein content of about 32 μg per seed (Figure 6B). By contrast, in an1 mutant seed, the inhibition on transcription factors and fatty acid synthesis genes are released, thus lipid and protein accumulation are elevated to about 45 and 36 μg per seed, respectively. Together with An2, a R2R3-MYB family protein, An1 could induce the expression of function genes (LAR and ANR) involved in PAs biosynthesis in WT tobacco seed coat. The accumulation of PAs in seed coat generates the black-seeded phenotype in WT seed. However, the PAs biosynthesis genes represent a lower expression level in mutant seeds and the PA accumulation is blocked, which lead to the yellow-seeded phenotype (Figure 9). PAs are widely observed in seed coat in a number of oilseed plants, such as Camelina sativa, Camellia oleifera, and Tree peony. We proposed that TT8 homolog genes could be used as an ideal target for enhancing seed lipid accumulation in these plants.
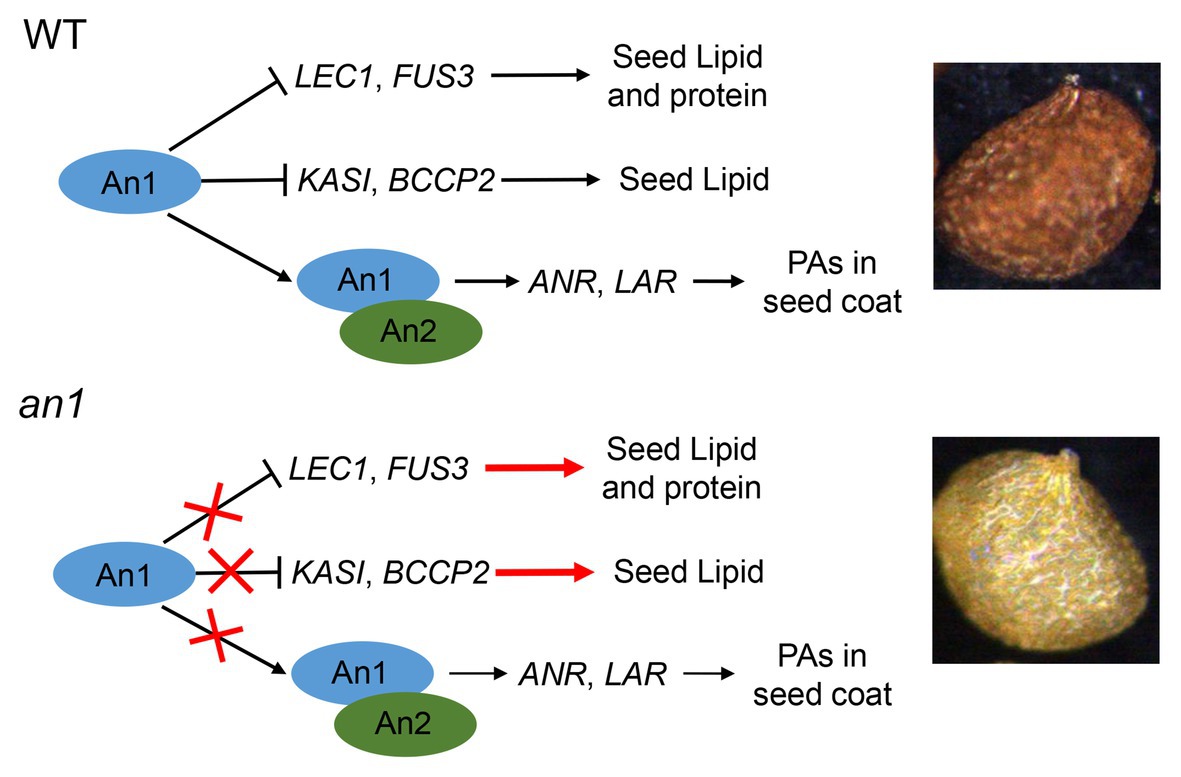
Figure 9. Model for An1-mediated regulation of PAs and lipid accumulation in tobacco seed. In WT tobacco seed, An1 protein could inhibit the expression of transcription factors (including LEC1 and FUS3) and fatty acid biosynthesis genes (including KASI and BCCP2). Together with An2, a R2R3-MYB family protein, An1 could induce the expression of function genes (LAR and ANR) involved in PA biosynthesis in WT tobacco seed coat. In an1 mutant seed, the inhibition on genes encoding transcription factor and fatty acid biosynthesis enzymes are released; thus, lipid and protein accumulation are enhanced in mutant lines. Compared with WT seed, the PA biosynthesis genes represent a lower expression level in mutant seeds and the PA accumulation is blocked, which lead to the yellow-seeded phenotype.
CRISPR-Cas9 System Could Be Used for de novo Tobacco Domestication for Biodiesel Feedstock Production
In recent years, the CRISPR-Cas9-based sequence-specific nucleases (SSNs) had been demonstrated to be the most simple and efficient tool for targeted gene editing. The CRISPR-Cas9 system has been successfully utilized in tobacco to generate the required mutagenesis for agronomic traits improvement and gene function characterization (Jiang et al., 2013; Gao et al., 2018; Schachtsiek and Stehle, 2019). Previous reports had demonstrated that homozygous mutant could be obtained in the first generation in diverse plants, including tomato, rice, grape, and poplar (Zhang et al., 2014; Fan et al., 2015; Pan et al., 2016; Wang et al., 2018). In this present paper, two homozygous mutant lines were generated in the first generation (Figure 2); our results demonstrated again that the CRISPR-Cas9 system is a highly efficient method for targeted gene mutation. The mutant lines showed an expected increase in seed lipid content (Figure 6A). In addition, the seed yield was not compromised (Figure 8).
Due to the ever-increasing global need for energy and environmental concerns about the effects of increasing carbon dioxide levels, the demand for biofuels has been dramatically increased in the past decades (Durrett et al., 2008). To meet the huge demand for biofuel feedstocks, de novo domestications of plants for biofuel production have attracted a lot of attention worldwide (Montes and Melchinger, 2016; Sainger et al., 2017). Tobacco seed oil is a promising feedstock for biodiesel production; however, current tobacco varieties are not bred for seed lipid production. With the development of CRISPR-Cas9 system, it has been proposed that CRISPR-Cas9-mediated genome editing could be used as a new tool by breeders to accelerate the domestication of semi-domesticated or even wild plants (Osterberg et al., 2017; Fernie and Yan, 2019; Khan et al., 2019). Low linoleic acid and high oleic acid content is a preferred character for biodiesel feedstock production, so we created a high-oleic acid tobacco variety using CRISPR-Cas9-mediated NtFAD2-2 gene editing technology in our previous work (Tian et al., 2020). In the past few years, CRISPR-Cas9-mediated domestication had been carried out in a number of wild plant species, such as wild tomato and groundcherry (Zsogon et al., 2017, 2018; Lemmon et al., 2018; Li et al., 2018).
Tobacco belongs to the Solanaceae family, which contains several well-characterized model crops, including tomato, potato, and pepper. Numerous regulators important for yield related traits, including fruit size, inflorescence, and shoot architecture, had been identified and characterized in these model crop species, especially tomato (Rothan et al., 2019). These genes include SP (SELF-PRUNING; Pnueli et al., 1998), fw2.2 (FRUIT WEIGHT 2.2; Frary et al., 2000), FASCIATED (Xu et al., 2015a), and MULTIFLORA (Lippman et al., 2008). Previous studies had demonstrated that the genetic regulation networks of agronomic traits were conserved in different plant taxa, which suggested that editing the homolog genes across species may generate similar phenotypes (Hussain et al., 2020). We suggested that the domestication knowledge from model crops could be translated into tobacco for the generation of high seed yield and high lipid content varieties. In this paper, seed lipid content was significantly increased by targeting a single gene and the multiplex CRISPR-Cas9 system could be applied in the future to simultaneously target several genes for multiple traits enhancement.
Conclusion
In this study, we showed that NtAn1a and NtAn1b genes were highly expressed in developing tobacco seed. Targeted mutation of NtAn1 genes were generated using the CRISPR-Cas9-mediated genome editing technology. Due to the defects in PAs biosynthesis, the mutant seeds showed the yellow-seeded phenotype. We showed that targeted mutagenesis of NtAn1 genes enhanced the seed lipid accumulation by about 18% than WT control seeds. The high knockout efficiency and significantly elevated lipid content in mutant seeds indicated that the CRISPR-Cas9 system could be applied to generate new tobacco varieties for biodiesel production in a faster way than traditional breeding method.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
YT and YX conceived and designed the study and wrote the manuscript. YT and XL performed the experiments. CF and YZ contributed in manuscript revision. TL, HQ, XL, and KC contributed to data acquisition. All authors reviewed, discussed, and interpreted the results. FC reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by the Special Project for New Transgenic Technologies and Methods of Ministry of Agriculture (no. 2016ZX08010001-010) and the Special Fund for Strategic Cooperation between Panzhihua and Sichuan University (no. 2018CDPZH-11). The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.599474/full#supplementary-material
Footnotes
References
Bai, Y., Pattanaik, S., Patra, B., Werkman, J. R., Xie, C. H., and Yuan, L. (2011). Flavonoid-related basic helix-loop-helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 234, 363–375. doi: 10.1007/s00425-011-1407-y
Banković-Ilić, I. B., Stamenković, O. S., and Veljković, V. B. (2012). Biodiesel production from non-edible plant oils. Renew. Sust. Energ. Rev. 16, 3621–3647. doi: 10.1016/j.rser.2012.03.002
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bell, J. M. (1993). Factors affecting the nutritional value of canola meal: a review. Can. J. Anim. Sci. 73, 689–697. doi: 10.4141/cjas93-075
Cai, J., Li, B., Chen, C., Wang, J., Zhao, M., and Zhang, K. (2016). Hydrothermal carbonization of tobacco stalk for fuel application. Bioresour. Technol. 220, 305–311. doi: 10.1016/j.biortech.2016.08.098
Carvalho, F. S., Fornasier, F., Leitão, J. O. M., Moraes, J. A. R., and Schneider, R. C. S. (2019). Life cycle assessment of biodiesel production from solaris seed tobacco. J. Clean. Prod. 230, 1085–1095. doi: 10.1016/j.jclepro.2019.05.177
Chen, M., Xuan, L., Wang, Z., Zhou, L., Li, Z., Du, X., et al. (2014). TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol. 165, 905–916. doi: 10.1104/pp.114.235507
Durrett, T. P., Benning, C., and Ohlrogge, J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. doi: 10.1111/j.1365-313X.2008.03442.x
Escaray, F. J., Passeri, V., Perea-Garcia, A., Antonelli, C. J., Damiani, F., Ruiz, O. A., et al. (2017). The R2R3-MYB TT2b and the bHLH TT8 genes are the major regulators of proanthocyanidin biosynthesis in the leaves of Lotus species. Planta 246, 243–261. doi: 10.1007/s00425-017-2696-6
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in populus in the first generation. Sci. Rep. 5:12217. doi: 10.1038/srep12217
Fernie, A. R., and Yan, J. (2019). De novo domestication: an alternative route toward new crops for the future. Mol. Plant 12, 615–631. doi: 10.1016/j.molp.2019.03.016
Frary, A., Nesbitt, T. C., Grandillo, S., Knaap, E., Cong, B., Liu, J., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. doi: 10.1126/science.289.5476.85
Gao, J., Tong, Z., Xu, B., Ling, J., Xiao, B., He, L., et al. (2018). CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 8 (CCD8) in tobacco affects shoot and root architecture. Int. J. Mol. Sci. 19:1062. doi: 10.3390/ijms19041062
Giannelos, P. N., Zannikos, F., Stournas, S., Lois, E., and Anastopoulos, G. (2002). Tobacco seed oil as an alternative diesel fuel: physical and chemical properties. Ind. Crops Prod. 16, 1–9. doi: 10.1016/S0926-6690(02)00002-X
Grisan, S., Polizzotto, R., Raiola, P., Cristiani, S., Ventura, F., di Lucia, F., et al. (2016). Alternative use of tobacco as a sustainable crop for seed oil, biofuel, and biomass. Agron. Sustain. Dev. 36:55. doi: 10.1007/s13593-016-0395-5
Heath, J. D., Weldon, R., Monnot, C., and Meinke, D. W. (1986). Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta 169, 304–312. doi: 10.2307/23378213
Hong, M., Hu, K., Tian, T., Li, X., Chen, L., Zhang, Y., et al. (2017). Transcriptomic analysis of seed coats in yellow-seeded Brassica napus reveals novel genes that influence proanthocyanidin biosynthesis. Front. Plant Sci. 8:1674. doi: 10.3389/fpls.2017.01674
Horsch, R. B. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Hussain, Q., Shi, J., Scheben, A., Zhan, J., Wang, X., Liu, G., et al. (2020). Genetic and signalling pathways of dry fruit size: targets for genome editing-based crop improvement. Plant Biotechnol. J. 18, 1124–1140. doi: 10.1111/pbi.13318
Jiang, J., Wang, Y., Xie, T., Rong, H., Li, A., Fang, Y., et al. (2015). Metabolic characteristics in meal of black rapeseed and yellow-seeded progeny of Brassica napus-Sinapis alba hybrids. Molecules 20, 21204–21213. doi: 10.3390/molecules201219761
Jiang, W., Zhou, H., Bi, H., Michael, F., Bing, Y., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 20:e188. doi: 10.1093/nar/gkt780
Khan, M. Z., Zaidi, S. S., Amin, I., and Mansoor, S. (2019). A CRISPR way for fast-forward crop domestication. Trends Plant Sci. 24, 293–296. doi: 10.1016/j.tplants.2019.01.011
Kumar, M., and Sharma, M. P. (2016). Selection of potential oils for biodiesel production. Renew. Sust. Energ. Rev. 56, 1129–1138. doi: 10.1016/j.rser.2015.12.032
Lemmon, Z. H., Reem, N. T., Dalrymple, J., Soyk, S., Swartwood, K. E., Rodriguez-Leal, D., et al. (2018). Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770. doi: 10.1038/s41477-018-0259-x
Lepiniec, L., Debeaujon, I., Routaboul, J. M., Baudry, A., Pourcel, L., Nesi, N., et al. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–430. doi: 10.1146/annurev.arplant.57.032905.105252
Li, Y., Beisson, F., Pollard, M., and Ohlrogge, J. (2006). Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67, 904–915. doi: 10.1016/j.phytochem.2006.02.015
Li, X., Chen, L., Hong, M., Zhang, Y., Zu, F., Wen, J., et al. (2012). A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PLoS One 7:e44145. doi: 10.1371/journal.pone.0044145
Li, P., Chen, B., Zhang, G., Chen, L., Dong, Q., Wen, J., et al. (2016). Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 210, 905–921. doi: 10.1111/nph.13816
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. doi: 10.1038/nbt.4273
Lim, S. H., Kim, D. H., Kim, J. K., Lee, J. Y., and Ha, S. H. (2017). A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front. Plant Sci. 8:1917. doi: 10.3389/fpls.2017.01917
Lippman, Z. B., Cohen, O., Alvarez, J. P., Abu-Abied, M., Pekker, I., Paran, I., et al. (2008). The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 6:e288. doi: 10.1371/journal.pbio.0060288
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., and Chen, L. L. (2017). CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Montes, J. M., and Melchinger, A. E. (2016). Domestication and breeding of Jatropha curcas L. Trends Plant Sci. 21, 1045–1057. doi: 10.1016/j.tplants.2016.08.008
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Osterberg, J. T., Xiang, W., Olsen, L. I., Edenbrandt, A. K., Vedel, S. E., Christiansen, A., et al. (2017). Accelerating the domestication of new crops: feasibility and approaches. Trends Plant Sci. 22, 373–384. doi: 10.1016/j.tplants.2017.01.004
Padmaja, L. K., Agarwal, P., Gupta, V., Mukhopadhyay, A., Sodhi, Y. S., Pental, D., et al. (2014). Natural mutations in two homoeologous TT8 genes control yellow seed coat trait in allotetraploid Brassica juncea (AABB). Theor. Appl. Genet. 127, 339–347. doi: 10.1007/s00122-013-2222-6
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6:24765. doi: 10.1038/srep24765
Pang, Y., Peel, G. J., Sharma, S. B., Tang, Y., and Dixon, R. A. (2008). A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc. Natl. Acad. Sci. U. S. A. 105, 14210–14215. doi: 10.1073/pnas.0805954105
Pnueli, L., Carmel-Goren, L., Hareven, D., Gutfinger, T., Alvarez, J., Ganal, M., et al. (1998). The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989. doi: 10.1016/0020-7462(90)90004-S
Poltronieri, P. (2016). “Chapter 6-Tobacco seed oil for biofuels” in Biotransformation of agricultural waste and by-products, 161–187.
Rahman, M., and Mcvetty, P. B. E. (2011). A review of Brassica seed color. Can. J. Plant Sci. 91, 437–446. doi: 10.4141/CJPS10124
Rothan, C., Diouf, I., and Causse, M. (2019). Trait discovery and editing in tomato. Plant J. 97, 73–90. doi: 10.1111/tpj.14152
Sainger, M., Jaiwal, A., Sainger, P. A., Chaudhary, D., Jaiwal, R., and Jaiwal, P. K. (2017). Advances in genetic improvement of Camelina sativa for biofuel and industrial bio-products. Renew. Sust. Energ. Rev. 68, 623–637. doi: 10.1016/j.rser.2016.10.023
Schaart, J. G., Dubos, C., Romero De La Fuente, I., van Houwelingen, A. M., de Vos, R. C., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Schachtsiek, J., and Stehle, F. (2019). Nicotine-free, nontransgenic tobacco (Nicotiana tabacum L.) edited by CRISPR-Cas9. Plant Biotechnol. J. 17, 2228–2230. doi: 10.1111/pbi.13193
Tanner, G. J., Francki, K. T., Abrahams, S., Watson, J. M., Larkin, P. J., and Ashton, A. R. (2003). Proanthocyanidin biosynthesis in plants. J. Biol. Chem. 278, 31647–31656. doi: 10.1074/jbc.M302783200
Tian, Y., Chen, K., Li, X., Zheng, Y., and Chen, F. (2020). Design of high-oleic tobacco (Nicotiana tabacum L.) seed oil by CRISPR-Cas9-mediated knockout of NtFAD2–2. BMC Plant Biol. 20:233. doi: 10.1186/s12870-020-02441-0
Usta, N., Aydoğan, B., Çon, A. H., Uğuzdoğan, E., and Özkal, S. G. (2011). Properties and quality verification of biodiesel produced from tobacco seed oil. Energy Convers. Manag. 52, 2031–2039. doi: 10.1016/j.enconman.2010.12.021
Veljkovic, V., Lakicevic, S., Stamenkovic, O., Todorovic, Z., and Lazic, M. (2006). Biodiesel production from tobacco (Nicotiana tabacum L.) seed oil with a high content of free fatty acids. Fuel 85, 2671–2675. doi: 10.1016/j.fuel.2006.04.015
Wang, Z., Chen, M., Chen, T., Xuan, L., Li, Z., Du, X., et al. (2014). TRANSPARENT TESTA2 regulates embryonic fatty acid biosynthesis by targeting FUSCA3 during the early developmental stage of Arabidopsis seeds. Plant J. 77, 757–769. doi: 10.1111/tpj.12426
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., Li, Z., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi: 10.1111/pbi.12832
Xie, D. Y., Sharma, S. B., Paiva, N. L., Ferreira, D., and Dixon, R. A. (2003). Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299, 396–399. doi: 10.1126/science.1078540
Xing, H. L., Dong, L., Wang, Z. -P., Zhang, H. -Y., Han, C. -Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Xu, W., Dubos, C., and Lepiniec, L. (2015b). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Xu, W., Grain, D., Bobet, S., Gourrierec, J. L., Thévenin, J., Kelemen, Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 202, 132–144. doi: 10.1111/nph.12620
Xu, C., Liberatore, K. L., MacAlister, C. A., Huang, Z., Chu, Y. H., Jiang, K., et al. (2015a). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792. doi: 10.1038/ng.3309
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2019). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18, 1153–1168. doi: 10.1111/pbi.13281
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. doi: 10.1111/pbi.12200
Zlatanov, M., Angelova, M., and Antova, G. (2007). Lipid composition of tobacco seeds. Bulgarian J. Agr. Sci. 38, 39–40.
Zsogon, A., Cermak, T., Naves, E. R., Notini, M. M., Edel, K. H., Weinl, S., et al. (2018). De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216. doi: 10.1038/nbt.4272
Keywords: tobacco, seed lipid, NtAn1, CRISPR-Cas9, biodiesel
Citation: Tian Y, Liu X, Fan C, Li T, Qin H, Li X, Chen K, Zheng Y, Chen F and Xu Y (2021) Enhancement of Tobacco (Nicotiana tabacum L.) Seed Lipid Content for Biodiesel Production by CRISPR-Cas9-Mediated Knockout of NtAn1. Front. Plant Sci. 11:599474. doi: 10.3389/fpls.2020.599474
Edited by:
Junhua Peng, Huazhi Rice Bio-Tech Co., Ltd., ChinaReviewed by:
Lingling Zhang, Chinese Academy of Sciences, ChinaJianhua Fan, East China University of Science and Technology, China
Copyright © 2021 Tian, Liu, Fan, Li, Qin, Li, Chen, Zheng, Chen and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Xu, xuying@scu.edu.cn
†These authors have contributed equally to this work
 Yinshuai Tian1,2,3†
Yinshuai Tian1,2,3† Tingting Li
Tingting Li Ying Xu
Ying Xu