Abstract
The bovine respiratory syncytial virus (BRSV) known as Bovine orthopneumovirus according to the international classification is one of the most important etiological agents of respiratory diseases in calves. At present, rapid and reliable methods to detect and measure the concentrations of this pathogen are needed. The objectives of the survey are developing the real-time polymerase chain reaction (PCR) to identify and quantify the BRSV RNA and, based on it, determining the number of the virus genomes in the respiratory tract of sick animals during the disease outbreaks. The nucleocapsid (N) protein gene of the virus served as the target for amplification. Messenger RNA (mRNA) of bovine GAPDH was used as a reference gene. A panel of positive control samples at known concentrations was used to estimate the virus and GAPDH numbers. The concentration of viral RNA extracted from the biomaterial samples was quantified relative to the bovine GAPDH mRNA level. The analytical sensitivity of PCR demonstrating high specificity and reproducibility was 1 × 103 genome equivalents per 1 cm3. All 273 samples of biological material taken from the animals with the respiratory diseases were analyzed. The virus genome was detected in 19.4% of samples. The viral RNA was more frequently detected in the lungs, which comprised 10.61% of positive samples. It was less frequently found in the mucous membranes of trachea and bronchi and the lymph nodes of the lungs, which comprised 0.73% of positive samples each. Concentrations of the virus in samples varied. The highest concentration was recorded in the lungs (1.3 ± 0.5—4.8 ± 0.47 log10 copies of BRSV/GAPDH RNA). The developed test kit may be used to quantify the concentration of the bovine respiratory syncytial virus in disease pathogenesis and to estimate the efficiency of vaccine or antivirus preparations for animals.
Similar content being viewed by others

INTRODUCTION
Respiratory diseases can cause economic losses in dairy farming, including from animal deaths, reduction in animal growth rates, and increase in costs for treatment, diagnoses, and preventive measures [1—3]. The respiratory syncytial virus (RSV) of cattle or bovine respiratory syncytial virus (BRSV) known as Bovine orthopneumovirus according to the international classification is one of the most important etiological agents of respiratory diseases. It is assigned to the family Pneumoviridae, the genus Orthopneumovirus [4]. The respiratory syncytial virus infection (RSVI) has been registered in all countries in the world countries in which livestock farming of an intensive type has been developed [5, 6].
The virus causes bronchiolitis, bronchitis, interstitial pneumonia, and emphysema in susceptible animals [5, 7].
The first information on identification of the bovine RSVI in Russia was reported in 1975 [8]. Many researchers have paid great attention to studying the specificity of disease distribution and developing the tools and methods for its diagnostics [9—12].
Detection of the epizootic situation for the bovine RSVI on large-scale dairy farms, especially farms involving the imported livestock, is considered an important task. Investigation of tropism of virus in the animal respiratory tract and its quantitative assessment is necessary. Such surveys are required to optimize antiepizootic measures and study the effectiveness of vaccines and antiviral preparations for animals [13—16].
It is well known that the bovine RSV is closely relative to the human RSV, which can cause clinical symptoms and pathological changes in calves similar to those in children [7, 16].
At present, there is no reliable model for the human RSV, while attempts to replicate the virus have been made with chimpanzees [16], cattle [17, 18], sheep [18], cotton rats [19], and mice [20, 21].
It has been proposed that cattle may represent an alternative model of human infection that can be used for both studying the final concentration of the virus in lungs of cotton rats or monkeys and assessing the dynamics of changes in the clinical symptoms and the concentrations of the virus at each stage of a disease [20].
Quantification of the bovine RSV accumulation in lungs is rather difficult, because of weak replication of the virus in cell cultures. The real-time polymerase chain reaction (PCR) test may be a promising approach. The accessible foreign literature reports some information on assessing the viral titers in nasopharyngeal swabs or bronchial–alveolar lavage fluids of the gnotobiont calves experimentally infected with the virus [22, 23] and evaluating the degree of pulmonary infection from the results of immunohistochemistry assays for lung tissues [24—26].
The concentration of pathogens weakly replicating in the cell cultures or easily destroyed may be measured with a quantitative PCR test. In addition, screening of the etalon genes, including 18S rRNA, GAPDH, ACTB, PRKG1, and TBP, [27], the expression of which may vary according to the types of tissues or the experimental conditions, is used to assess the integrity of nucleic acids.
At present, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene is used as an etalon (positive control) for assessing the gene expression with quantitative PCR tests. The levels of GAPDH mRNA in different organs of cattle are rather stable [27]. Therefore, it was used in quantitative evaluation of the bovine RSV RNA.
The objectives of the survey are to develop a real-time PCR test to identify and quantify BRSV RNA and, on this basis, determining the number of the virus genomes in the respiratory tract of sick animals during disease outbreaks.
EXPERIMENTAL
Surveys were carried out at large-scale dairy farms (megafarms) during outbreaks of mass infectious respiratory illnesses in animals. Overall, 273 random samples of biomaterial taken from calves aged 10 days to 6 months were analyzed. They included 171 and 45 samples taken from the lungs and nasal swabs, respectively; 24 samples collected from exudates in the trachea, bronchi, and nasal sinuses; 14 samples of bronchial and tracheal mucosa; and 10 and 9 samples collected in the lymph nodes and bronchi, respectively, of animals that had died or were slaughtered for postmortem diagnostic tests. The samples in freezing or transport media were delivered to the laboratory. The delivery time was no longer than 12 h from the moment of sampling. The samples were stored at –80°C.
The samples were grinned with sterile sand in a porcelain mortar to prepare 10% saline suspensions, which were centrifuged at 3000 rpm for 15 min and analyzed in the RT-PCR. Prior to centrifuging, the samples of thick nasal swabs and exudates were diluted with the physiological solution in a proportion of 1 : 5. In order to isolate RNA, supernatant of 100 μL each was used.
Bovine RSV Strain RSB obtained at the Vyshelessky Belarusian Research Institute of Experimental Veterinary Science was used. The viral concentration comprising 103.5 \({\text{TC}}{{{\text{D}}}_{{{\text{50}}}}}\)/cm3 was measured with the technique of titration in the calf lung culture initially trypsinized.
RNA Extraction and Reverse Transcription
RNA was isolated from the organ or a nasal-swab homogenate of 100 μL with a Litekh kit (Russia) according to the manufacturer’s recommendations. The purified RNA was resuspended in 50 μL of RNA buffer. To perform the reverse transcription, 10 μL of isolated RNA was used. The reaction was performed with the Reverta-L kit (Russia) according to the manufacturer’s recommendations. After reaction, the sample volume comprised 40 μL.
Primers and Probes
Primers and probes were selected with nucleotide sequences obtained at the GenBank of the International Nucleotide Sequence Database. The Vector NTI 9.0 software (Informax, United States) and the database online resources of the National Center for Biotechnology Information (NCBI) were used to align the obtained sequences and analyze and estimate primers and probes for studying their specificity. The primers and the probes to the mRNA of bovine gene GAPDH were used as the internal control (Table 1).
Setting Up PCR Reactions
The PCR reaction was performed in a 30 μL mixture containing 5 μL kDNA, 10 pMol each of primer and probe, and a ready mixture of BioMaster real-time PCR reagents (Biolabmix, Russia). An amplification program with a cycle of 95°C for 5 min, followed by 45 cycles of 95°C for 15 min, and a cycle of 60°C for 1 min was used.
Positive Control Samples
The positive control samples were produced through cloning synthesized DNA fragments carrying the virus-specific insert into the PCR 2.1 plasmid (Invitrogen, United States) to transform the TOP 10 Escherichia coli cells (Invitrogen).
DNA Concentration Measurement
To measure the plasmid DNA concentration, a Quant-iT DNA HS Assay Kit (Invitrogen) and Quibit Fluorometer (Invitrogen) were used.
Quantitative Assessment of Viral RNA
To quantify RNA in the samples, the standard linear curve of amplification in a series of tenfold dilutions for glycoprotein N and GAPDH genes at known concentrations was constructed. The RNA virus and GAPDH numbers in the sample were assessed with a method of comparison of sample Ct values to a standard curve and expressed in log10 copies of RNA virus per 105 copies of GAPDH (log10 BRSV/GAPDH) [15].
Verification of Results
To verify the results, the biomaterial samples were analyzed with a PCR-based testing system for diagnostics of bovine RSVI that was developed at the Institute of Experimental Veterinary Science of Siberia and the Far East, Siberian Federal Science Centre for Agro-BioTechnologies, Russian Academy of Sciences, under state registration certificate no. PVR-1-8.9/02499. This testing system was developed to identify the fragment of the virus F glycoprotein gene.
RESULTS AND DISCUSSION
Table 1 shows the nucleotide sequences of primers and probes. The specificity of the selected oligonucleotides was analyzed at the estimation stage with the nucleotide Blast NCBI software. The survey outcomes may prove that the selected primers and probes had no homology to bovine DNA and the most common viruses involved in the etiology of respiratory diseases. The indicated viral group includes the viruses of rhonotracheitis (Bovine herpes virus type 1 and BoHV-1), viral diarrhea viruses (Bovine pestivirus type A, B, and H, (BVDV)), coronoviruses (Bovine coronavirus and BoCV), rhinoviruses (Bovine Rhinovirus and BoRV), and parainfluenza-3 viruses (Bovine parainfluenza type 3 virus and PI-3).
Positive control samples (PCSs) were used to assess the efficiency of the real-time PCR, which required them to be successively diluted tenfold. The surveys were carried out in three replicates. Diluted PCS of 5 μL each per reaction were added to the PCR mix. The sensitivity of the PCR amplification for the N PCB bovine nucleoprotein gene was 2.5 × 103 genome equivalents per 1 cm3. The GAPDH gene comprised 1.2 × 103 genome equivalents per 1 cm3. Linear dependences R 2 in high-dimensional genomic data came to 0.9873 and 0.9976, respectively. The coefficient of reproducibility for different dilutions varied from 0.32 to 3.57%, which can indicate a high reaction efficiency (Table 2).
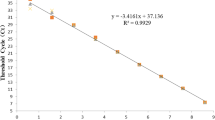
The efficiency of a reaction in quantification of the viral RNA concentration was assessed by testing the tenfold dilutions of the RSV PCB control strain bovine with the concentration comprising 103.5 \({\text{TC}}{{{\text{D}}}_{{{\text{50}}}}}\)/cm3 that was previously measured. Therefore, seven successive tenfold dilutions of the virus were prepared. RNA was isolated from them and analyzed with the PCR test. The values were compared with the curve data on the PSO dilutions. The threshold of the reaction sensitivity comprised 100.5 \({\text{TC}}{{{\text{D}}}_{{{\text{50}}}}}\)/cm3 (Fig. 1), which matches 5 log10 PCS/cm3.
The developed PCR test was used for analyzing 273 samples of biomaterial taken from the animals. The virus was detected in 53 samples, which comprised 19.4% (Table 3).
Table 3 shows that the virus genome was more frequently found in the samples collected from lungs, tracheal and bronchial exudates, nasal swabs, and bronchial sputum, which were 10.61, 4.03, 2.2, and 1.1% positive samples, respectively. The virus was found less frequently in the tracheal and bronchial mucus and lung lymph nodes, comprising 0.73% positive samples each. The virus concentration in the biomaterial samples varied, which can indicate different stages of infection in animals at sampling. The highest concentrations of the virus genome were measured in the lungs (1.3 ± 0.5–4.8 ± 0.47 log10 RNA copies BRSV/GAPDH), the nasal swabs (1.5 ± 0.75—2.1 ± 0.25 log10 RNA copies BRSV/GAPDH), and the exudates of trachea, bronchi, and nasal sinuses (0.3 ± 0.21—2.8 ± 0.15 log10 RNA copies BRSV/GAPDH).
The results coincided 100% with the data produced in analyzing the same biomaterial samples with the previously developed testing system based on the electrophoretic detection of amplification products.
The gene of the envelope glycoprotein of virus N was chosen to develop a real-time PCR kit allowing us to identify and quantify bovine RSV RNA in the samples of animal biomaterial, since it is one of the most conservative genes. Thus, mRNA of gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a gene for comparing and regulating the PCR, since it is most stably expressed at a high level in the bovine lungs. Both reactions possessed high sensitivity and reproducibility. The coefficient of reproducibility for different dilutions varied in the range of 0.32–3.57%, which may indicate a high efficiency of a reaction (Table 2).
The diagnostic sensitivity of a reaction was determined with testing the tenfold dilutions of the RSV PCB bovine control strain with the known concentration. The virus concentration of 103.5 \({\text{TC}}{{{\text{D}}}_{{{\text{50}}}}}\)/cm3 matched 8 log10 PCS, while the concentration of 100.5 \({\text{TC}}{{{\text{D}}}_{{{\text{50}}}}}\)/cm3 matched 5 log10 PCS. The obtained results generally coincide with data of foreign researchers concerning the high sensitivity of the quantitative PCR, comprising 0.1 \({\text{TC}}{{{\text{D}}}_{{{\text{50}}}}}\)/cm3 in testing the vaccine virus and the samples taken from the transtracheal wash of the sick calves [28, 29].
The distribution of the virus in the respiratory tract of calves infected with RSVI was previously studied with the PCR test in the electrophoresis format. However, the method failed to measure the agent concentration because of its limitations [30].
In this work, quantitative real-time PCR was performed to make an attempt to quantify the virus genome in random samples taken from sick calves without taking into consideration the stage of spread of infection.
In this survey, the maximum accumulation of the virus was recorded in the lungs and nasal swabs, which may confirm the data on viral tropism to the interstitial tissue of the lungs [5, 7]. The virus concentration in bronchi was lower than that in the lungs, which may be associated with the agent migrating from the mucous membrane to the lung tissue and changes caused by tissue destruction. The interstitial pneumonia and emphysema in the lungs in the calves used for postmortem testing may serve as indirect evidence of this.
CONCLUSIONS
A survey for developing a kit of components to identify and quantify bovine RSV RNA in samples of biomaterial taken from animals naturally infected during an infection outbreak has been carried out. The outcomes for determining the diagnostic and analytical sensitivity of the kit have proved that it possesses high specificity and sensitivity. Therefore, it may be used to identify and quantify bovine RSV when studying the disease pathogenesis and the efficiency of vaccines or antiviral preparations.
REFERENCES
Grandin, T., Welfare problems in cattle, pigs, and sheep that persist even though scientific research clearly shows how to prevent them, Animals, 2018, vol. 8, p. 124. https://doi.org/10.3390/ani8070124
Delabouglise, A., James, A., Valarcher, J.F., Hagglünd, S., Raboisson, D., and Rushton, J., Linking disease epidemiology and livestock productivity: The case of bovine respiratory disease in France, PLoS One, 2017, vol. 12, no. 12, p. e0189090. https://doi.org/10.1371/journal.pone.0189090
Shabunin, S.V., Shakhov, A.G., Chernitskii, A.E., Zolotarev, A.I., and Retskii, M.I., Respiratory diseases of calves: a modern approach to the problem, Veterinariya, 2015, no. 5, pp. 3–13. https://doi.org/10.17116/rosakush20151514-8
Virus Taxonomy: 2018 Release, International Committee on Taxonomy of Viruses. https://talk.ictvonline.org/.
Glotov, A.G., Petrova, O.G., Glotova, T.I., Nefedchenko, A.V., Tatarchuk, A.T., Koteneva, S.V., et al., The spread of viral respiratory diseases of cattle, Veterinariya, 2002, no. 3, pp. 17–21.
Taylor, J.D., Fulton, R.W., Lehenbauer, T.W., Step, D.L., and Confer, A.W., The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors?, Can. Vet. J., 2010, vol. 51, pp. 1095–1102. PMID 21197200
Valarcher, J.-F. and Taylor, G., Bovine respiratory syncytial virus infection, Vet. Res., 2007, vol. 38, pp. 153–180. https://doi.org/10.1051/vetres:2006053
Gunenkov, V.V., Khalenev, G.A., and Syurin, V.N., Respiratory syncytial viral infection, Zhivotnovod. Vet., 1975, vol. 8, pp. 70–76.
Glotova, T.I., Kungurtseva, O.V., Glotov, A.G., Stroganova, I.Ya., Voitova, K.V., and Gal’nbek, T.V., Isolation and typing of respiratory syncytial virus in cattle using RT-PCR, Sib. Vestn. S-kh. Nauki, 2010, vol. 10, pp. 59–64.
Glotov, A.G., Glotova, T.I., Koteneva, S.V., Nefedchenko, A.V., and Voitova, K.V., Features of the epizootic situation on respiratory syncytial infection of cattle in farms for the production of milk, Veterinariya, 2010, no. 7, pp. 21–25.
Glotov, A.G., Glotova, T.I., and Stroganova, I.Ya., Detection of bovine respiratory syncytial virus using RT-PCR, Vopr. Virusol., 2011, vol. 56, no. 5, pp. 34–37.
Zhuravleva, E.A., Nosoarea of bovine respiratory syncytial virus infection, Veterinariya, 2018, no. 12, pp. 3–8. https://doi.org/10.30896/0042-4846.2018.21.12.03-08
Jordan, R., Shao, M., Mackman, R.L., Perron, M., Cihlar, T., Lewis, S.A., et al., Antiviral efficacy of a respiratory syncytial virus (RSV) fusion inhibitor in a bovine model of RSV infection, Antimicrob. Agents Chemother., 2015, vol. 59, pp. 4889–4900. https://doi.org/10.1128/AAC.00487-15
Meyer, G., Deplanche, M., and Schelcher, F., Human and bovine respiratory syncytial virus vaccine research and development, Comp. Immunol. Microbiol. Infect. Dis., 2008, vol. 31, nos. 2–3, pp. 191–225. https://doi.org/10.1016/j.cimid.2007.07.008
Aranda, S.S. and Polack, F.P., Prevention of pediatric respiratory syncytial virus lower respiratory tract illness: perspectives for the next decade, Front. Immunol., 2019, vol. 10, p. 1006. https://doi.org/10.3389/fimmu.2019.01006
Kirolos, A., Christides, A., Xian, S., Reeves, R., Nair, H., and Campbell, H., A landscape review of the published research output relating to respiratory syncytial virus (RSV) in North & Central America and Europe between 2011–2015, J. Global Health, 2019, vol. 9, no. 1, p. 010425. https://doi.org/10.7189/jogh.09.010425
Gershwin, L.J., Schelegle, E.S., Gunther, R.A., Anderson, M.L., Woolums, A.R., Larochelle, D.R., et al., A bovine model of vaccine enhanced respiratory syncytial virus pathophysiology, Vaccine, 1998, vol. 16, pp. 1225–1236. https://doi.org/10.1016/S0264-410X(98)80123-0
Masot, A.J., Gomez-Tejedor, C., Gazquez, A., and Redondo, E., Pathological study of experimentally induced bovine respiratory syncytial virus infection in lambs, Zentralbl. Veterinaermed., Reihe B, 1996, vol. 43, pp. 233–243.
Boukhvalova, M.S., Prince, G.A., and Blanco, J.C., The cotton rat model of respiratory viral infections, Biologicals, 2009, vol. 37, pp. 152–159. https://doi.org/10.1016/j.biologicals.2009.02.017
Almeida, R.S., Domingues, H.G., Coswig, L.T., d’Arce, R.C.F., de Carvalho, R.F., and Arns, C.W., Detection of bovine respiratory syncytial virus in experimentally infected balb/c mice, Vet. Res., 2004, vol. 35, pp. 189–197.
Rudd, P.A., Chen, W., and Mahalingam, S., Mouse and cotton rat models of human respiratory syncytial virus, Methods Mol. Biol., 2016, vol. 1442, pp. 209–217. https://doi.org/10.1007/978-1-4939-3687-8_15
Boxus, M., Letellier, C., and Kerkhofs, P., Real Time RT-PCR for the detection and quantitation of bovine respiratory syncytial virus, J. Virol. Methods, 2005, vol. 125, no. 2, pp. 125–130. https://doi.org/10.1016/j.jviromet.2005.01.008
Kimman, T.G., Zimmer, G.M., Straver, P.J., and de Leeuw, P.W., Diagnosis of bovine respiratory syncytial virus infections improved by virus detection in lung lavagesamples, Am. J. Vet. Res., 1986, vol. 47, no. 1, pp. 143–147.
Yaman, T., Büyükbayram, H., Özyıldız, Z., Terzi, F., Uyar, A., Keles, Ö.F., et al., Detection of bovine respiratory syncytial virus, Pasteurella Multocida, and Mannheimia Haemolytica by immunohistochemical method in naturally-infected cattle, J. Vet. Res., 2018, vol. 62, no. 4, pp. 439–445. https://doi.org/10.2478/jvetres-2018-0070
Klem, T.B., Sjurseth, S.K., Sviland, S., Gjerset, B., Myrmel, M., and Stokstad, M., Bovine respiratory syncytial virus in experimentally exposed and rechallenged calves; viral shedding related to clinical signs and the potential for transmission, BMC Vet. Res., 2019, vol. 15, no. 1, p. 156. https://doi.org/10.1186/s12917-019-1911-z
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al., Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes, Genome Biol., 2002, vol. 3, no. 7, 0034.1–0034.11.
Zhao, H., Liu, J., Li, Y., Yang, C., Zhao, S., Liu, J., et al., Validation of reference genes for quantitative Real-Time PCR in bovine PBMCs transformed and non-transformed by Theileria annulate, Korean J. Parasitol., 2016, vol. 54, no. 1, pp. 39–46. https://doi.org/10.3347/kjp.2016.54.1.39
Timsit, E., Maingourd, C., Le Dréan, E., Belloc, C., Seegers, H., Douart, A., et al., Evaluation of a commercial real-time reverse transcription polymerase chain reaction kit for the diagnosis of Bovine respiratory syncytial virus infection, J. Vet. Diagn. Invest., 2010, vol. 22, no. 2, pp. 238–241. https://doi.org/10.1177/104063871002200211
Timsit, E., Le Dréan, E., Maingourd, C., Belloc, C., Guattéo, R., Bareille, N., et al., Detection by real-time RT-PCR of a bovine respiratory syncytial virus vaccine in calves vaccinated intranasally, Vet. Rec., 2009, vol. 165, no. 8, pp. 230–233. https://doi.org/10.1136/vr.165.8.230
Koteneva, S.V., Voitova, K.V., Glotova, T.I., Stroganova, I.Ya., and Glotov, A.G., The frequency of detection of the genome of the respiratory syncytial virus in cattle during outbreaks of bronchopneumonia in dairy complexes, Ross. Vet. Zh. S-kh. Zhivotn., 2016, no. 3, pp. 18–21.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
STATEMENT ON THE WELFARE OF ANIMALS
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The article does not contain any research using animals as subjects.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Additional information
Translated by O. Zhiryakova
ADDITIONAL INFORMATION
Nefedchenko A.V., https://orcid.org/0000-0002-4181-4268; e-mail: nav-vet@mail.ru
Glotov A.G., https://orcid.org/0000-0002-2006-0196; e-mail: glotov_vet@mail.ru
Koteneva S.V., https://orcid.org/0000-0003-2649-7505; e-mail: t-glotova@mail.ru
Glotova T.I., https://orcid.org/0000-0003-3538-8749; e-mail: koteneva-sv@mail.ru
About this article
Cite this article
Nefedchenko, A.V., Glotov, A.G., Koteneva, S.V. et al. Developing and Testing a Real-Time Polymerase Chain Reaction to Identify and Quantify Bovine Respiratory Syncytial Viruses. Mol. Genet. Microbiol. Virol. 35, 168–173 (2020). https://doi.org/10.3103/S0891416820030052
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0891416820030052