Abstract
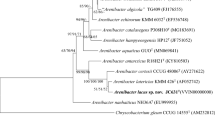
A yellow-colored, Gram-stain-positive, rod shaped, non-motile bacterium, designated as strain JC619T, was isolated from the sediment of Chilika lagoon, India. Strain JC619T shows highest 16S rRNA gene sequence similarity (99.08%) with Isoptericola chiayiensis KCTC 19740T followed by Isoptericola halotolerans KCTC 19046T (98.6%) and other members of the genus Isoptericola (< 98%). NaCl is required for growth of strain JC619T and tolerates up to 18% (w/v) and pH up to 10. Strain JC619T grows optimally at temperature 30 °C, NaCl concentration of 3% (w/v), and at pH 7.5. The genome size of strain JC619T is 3.2 Mb with G+C content of 73.0 mol%. ANI scores of strain JC619T are 81.9% and 80.1% and dDDH values are 24.4% and 22.7% with I. chiayiensis KCTC 19740T and I. halotolerans KCTC 19046T, respectively. Respiratory quinones are MK-9(H4) and MK-9(H2). Predominant fatty acids (> 10%) are anteiso-C15:0, iso-C16:0, and iso-C15:0. Major polar lipids of strain JC619T are phosphatidylglycerol, phosphatidylinositolmannoside, diphosphatidylglycerol, and phosphatidylinositol. Strain JC619T is catalase positive but cytochrome oxidase negative and reduces nitrate. The genomic distinction of strain JC619T with its nearest related species of the genus Isoptericola is well supported with chemotaxonomic characteristics and differential physiological properties. Therefore, strain JC619T represents a new species under the genus Isoptericola for which Isoptericola sediminis sp. nov. is proposed. Type strain is JC619T (=KCTC 49244T =NBRC 114063T).


Similar content being viewed by others
References
Stackebrandt E, Schumann P (2004) Reclassification of Cellulosimicrobium variabile Bakalidou et al. 2002 as Isoptericola variabilis gen. nov., comb. nov. Int J Syst Evol Microbiol 54:685–688
Bakalidou A, Kämpfer P, Berchtold M, Kuhnigk T, Wenzel M et al (2002) Cellulosimicrobium variabile sp. nov., a cellulolytic bacterium from the hindgut of the termite Mastotermes darwiniensis. Int J Syst Evol Microbiol 52:1185–1192
Schumann P, Weiss N, Stackebrandt E (2001) Reclassification of Cellulomonas cellulans (Stackebrandt and Keddie 1986) as Cellulosimicrobium cellulans gen. nov., comb. nov. Int J Syst Evol Microbiol 51:1007–1010
Stackebrandt E, Rainey FA, Ward-Rainey NL (1997) Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 47:479–491
Ming H, Niu M-M, Cheng L-J, Zhang Y-M, Yi B-F, Xia T-T, Li M, Nie G-X (2020) Isoptericola halalbus sp. nov., a halotolerant actinobacterium isolated from saline lake sediment. Int J Syst Evol Microbiol 70:4661–4667
Wu Y, Li WJ, Tian W, Zhang LP, Xu L et al (2010) Isoptericola jiangsuensis sp. nov., a chitin-degrading bacterium. Int J Syst Evol Microbiol 60:904–908
Huang Z, Sheng XF, Zhao F, He LY, Huang J, Wang Q (2012) Isoptericola nanjingensis sp. nov., a mineral-weathering bacterium. Int J Syst Evol Microbiol 62:971–976
Mohapatra A, Mohanty RK, Mohanty SK, Bhatta KS, Das NR (2007) Fisheries enhancement and biodiversity assessment of fish, prawn and mud crab in Chilika lagoon through hydrological intervention. Wetl Ecol Manag 15:229–251
Rath J, Adhikary SP (2008) Biodiversity assessment of algae in Chilika Lake, east coast of India. In: Mohanty PK (ed) Monitoring and modelling lakes and coastal environments. Springer, Dordrecht, pp 22–33
Kumar D, Gaurav K, Jagadeeshwari U, Deepshikha G, Sasikala Ch, Ramana ChV (2020) Roseimaritima sediminicola sp. nov., a new member of Planctomycetaceae isolated from Chilika lagoon. Int J Syst Evol Microbiol 70:2616–2623
Kumar D, Gaurav K, Sreya PK, Shabbir A, Jagadeeshwari U, Sasikala Ch, Ramana ChV (2020) Gimesia chilikensis sp. nov., a haloalkali-tolerant planctomycete isolated from Chilika lagoon and emended description of the genus Gimesia. Int J Syst Evol Microbiol 70:3647–3655
Kumar D, Kumar G, Uppada J, Ahmed S, Sasikala C, Ramana CV (2020c) Descriptions of Roseiconus nitratireducens gen. nov. sp. nov. and Roseiconus lacunae sp. nov. Arch Microbiol. https://doi.org/10.1007/s00203-020-02078-5
Lakshmi KV, Sasikala C, Takaichi S, Ramana C (2011) Phaeospirillum oryzae sp. nov., a spheroplast-forming, phototrophic alphaproteobacterium from a paddy soil. Int J Syst Evol Microbiol 61:1656–1661
Beumer A, Robinson JB (2005) A broad-host-range, generalized transducing phage (SN-T) acquires 16S rRNA genes from different genera of bacteria. Appl Environ Microbiol 71:8301–8304
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Meier-Kolthoff JP, Klenk HP, Göker M (2014) Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age. Int J Syst Evol Microbiol 64:352–356
Rodriguez RLM, Konstantinidis KT (2014) Bypassing cultivation to identify bacterial species. Microbe 9:111–118
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Na SI, Kim YO, Yoon SH, Ha SM, Baek I et al (2018) UBCG: up-to- date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Smibert RM, Krieg NR (1981) General characterization. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB (eds) Manual of methods for general microbiology. American Society for Microbiology, Washington DC, pp 409–443
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical note 101
Kates M (1986) Techniques of lipidology. Isolation, analysis and identification of lipids. Lab Tech Biochem Mol Biol 3:100–112
Oren A, Duker S, Ritter S (1996) The polar lipid composition of Walsby’s square bacterium. FEMS Microbiol Lett 138:135–140
Imhoff JF (1984) Quinones of phototrophic purple bacteria. FEMS Microbiol Lett 25:85–89
Hiraishi A, Hoshino Y, Kitamura H (1984) Isoprenoid quinone composition in the classification of Rhodospirillaceae. J Gen Appl Microbiol 30:197–210
Huang Z, Sheng XF, Zhao F, He LY, Huang J et al (2012) Isoptericola nanjingensis sp. nov., a mineral-weathering bacterium. Int J Syst Evol Microbiol 62:971–976
Chun J, Oren A, VentosaA CH, Arahal DR et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Groth I, Schumann P, Schütze B, Gonzalez JM, Laiz L et al (2005) Isoptericola hypogeus sp. nov., isolated from the Roman catacomb of Domitilla. Int J Syst Evol Microbiol 55:1715–1719
Yoon JH, Schumann P, Kang SJ, Jung SY, Oh TK (2006) Isoptericola dokdonensis sp. nov., isolated from soil. Int J Syst Evol Microbiol 56:2893–2907
Kämpfer P, Glaeser SP, Kloepper JW, Hu CH, McInroy JA (2016) Isoptericola cucumis sp. nov., isolated from the root tissue of cucumber (Cucumis sativus). Int J Syst Evol Microbiol 66:2784–2788
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, D., U., J., A., K. et al. Isoptericola sediminis sp. nov., Isolated from Chilika Lagoon. Curr Microbiol 78, 848–855 (2021). https://doi.org/10.1007/s00284-020-02325-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02325-4