Genome-Wide Association Study of Natural Variation in Arabidopsis Exposed to Acid Mine Drainage Toxicity and Validation of Associated Genes with Reverse Genetics
Abstract
:1. Introduction
2. Results
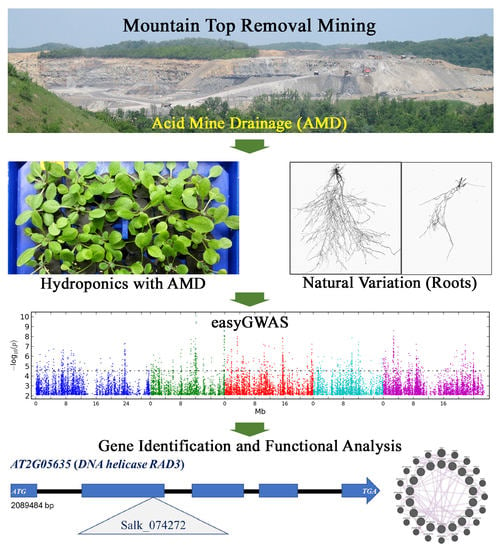
2.1. AMD Analysis and Hydroponic Growth System
2.2. Natural Variation in Root Growth Response to Acidity
2.3. GWAS by Using easyGWAS
2.4. Candidate Genes Associated with Adaptation to AMD
2.5. Subcellular Location and Protein Interaction Network during Acidity Tolerance
2.6. miR399b and DNA helicase RAD3 Knockout Mutants Show Tolerance to AMD Toxicity
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Collection of AMD and Analysis for Metals
4.2. Hydroponic Culture Growth Conditions
4.3. Documentation of Phenotypic Traits and Statistical Analysis
4.4. GWAS
4.5. Gene Ontology Enrichment and Coexpression Network Analysis
4.6. Genotyping of T-DNA Mutants and Phenotypic Validation
4.7. Complementary DNA (cDNA) Synthesis and RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wuana, R.A.; Okieimen, F.E. Heavy metals in contaminated soils: A review of sources, chemistry, risks and best available strategies for remediation. Isrn Ecol. 2011, 2011. [Google Scholar] [CrossRef] [Green Version]
- Palmer, M.A.; Bernhardt, E.S.; Schlesinger, W.H.; Eshleman, K.N.; Foufoula-Georgiou, E.; Hendryx, M.S.; Lemly, A.D.; Likens, G.E.; Loucks, O.L.; Power, M.E. Mountaintop mining consequences. Science 2010, 327, 148–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eia, U. Annual Energy Outlook 2015: With Projections to 2040; U.S. Energy Information Administration (EIA): Washington, DC, USA, 2017.
- Akcil, A.; Koldas, S. Acid Mine Drainage (AMD): Causes, treatment and case studies. J. Clean. Prod. 2006, 14, 1139–1145. [Google Scholar] [CrossRef]
- Cook, A.M.; Fritz, S.J. Environmental impacts of acid leachate derived from coal-storage piles upon groundwater. Water Air Soil Pollut. 2002, 135, 371–388. [Google Scholar] [CrossRef]
- Bernhardt, E.S.; Palmer, M.A. The environmental costs of mountaintop mining valley fill operations for aquatic ecosystems of the Central Appalachians. Ann. N. Y. Acad. Sci. 2011, 1223, 39–57. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V.; Hoekenga, O.A.; Pineros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef]
- Van Assche, F.; Clijsters, H. Effects of metals on enzyme activity in plants. Plant Cell Environ. 1990, 13, 195–206. [Google Scholar] [CrossRef]
- Koyama, H.; Toda, T.; Hara, T. Brief exposure to low-pH stress causes irreversible damage to the growing root in Arabidopsis thaliana: Pectin–Ca interaction may play an important role in proton rhizotoxicity. J. Exp. Bot. 2001, 52, 361–368. [Google Scholar] [PubMed]
- Jennings, S.; Neuman, D.; Blicker, P. Acid Mine Drainage and Effects on Fish Health and Ecology: A Review Reclamation Research Group; Reclamation Research Group Publication: Bozeman, MT, Canada, 2008. [Google Scholar]
- Soucek, D.J.; Cherry, D.S.; Currie, R.J.; Latimer, H.A.; Trent, G.C. Laboratory to field validation in an integrative assessment of an acid mine drainage–impacted watershed. Environ. Toxicol. Chem. Int. J. 2000, 19, 1036–1043. [Google Scholar]
- Hall, J.Á. Cellular mechanisms for heavy metal detoxification and tolerance. J. Exp. Bot. 2002, 53, 1–11. [Google Scholar] [CrossRef]
- Fukuda, T.; Saito, A.; Wasaki, J.; Shinano, T.; Osaki, M. Metabolic alterations proposed by proteome in rice roots grown under low P and high Al concentration under low pH. Plant Sci. 2007, 172, 1157–1165. [Google Scholar] [CrossRef] [Green Version]
- Hede, A.; Skovmand, B.; Lopez Cesati, J. Acid Soils and Aluminum Toxicity. Application of Physiology in Wheat Breeding; 9706480773; Centro Internacional de Mejoramiento de Maíz y Trigo, (CIMMYT): México, Mexico, 2001. [Google Scholar]
- Martínez-Camacho, J.L.; González-de La Vara, L.; Hamabata, A.; Mora-Escobedo, R.; Calderón-Salinas, V. A pH-stating mechanism in isolated wheat (Triticum aestivum) aleurone layers involves malic acid transport. J. Plant Physiol. 2004, 161, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Zheng, S.J.; He, Y.F.; Matsumoto, H. Aluminium resistance requires resistance to acid stress: A case study with spinach that exudes oxalate rapidly when exposed to Al stress. J. Exp. Bot. 2005, 56, 1197–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Saminathan, T.; Malkaram, S.A.; Patel, D.; Taylor, K.; Hass, A.; Nimmakayala, P.; Huber, D.H.; Reddy, U.K. Transcriptome analysis of invasive plants in response to mineral toxicity of reclaimed coal-mine soil in the appalachian region. Environ. Sci. Technol. 2015, 49, 10320–10329. [Google Scholar] [CrossRef] [PubMed]
- Foy, C.D. Plant adaptation to acid, aluminum-toxic soils. Commun. Soil Sci. Plant Anal. 1988, 19, 959–987. [Google Scholar] [CrossRef]
- Huang, C.-F.; Yamaji, N.; Ma, J.F. Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 2010, 153, 1669–1677. [Google Scholar] [CrossRef] [Green Version]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Díaz, A.; Brito-Argáez, L.; Munnik, T.; Hernández-Sotomayor, S.T. Aluminum inhibits phosphatidic acid formation by blocking the phospholipase C pathway. Planta 2007, 225, 393–401. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kobayashi, Y.; Watanabe, T.; Shaff, J.E.; Ohta, H.; Kochian, L.V.; Wagatsuma, T.; Kinraide, T.B.; Koyama, H. Molecular and physiological analysis of Al3+ and H+ rhizotoxicities at moderately acidic conditions. Plant Physiol. 2013, 163, 180–192. [Google Scholar] [CrossRef] [Green Version]
- Lager, I.; Andréasson, O.; Dunbar, T.L.; Andreasson, E.; Escobar, M.A.; Rasmusson, A.G. Changes in external pH rapidly alter plant gene expression and modulate auxin and elicitor responses. Plant Cell Environ. 2010, 33, 1513–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bose, J.; Babourina, O.; Shabala, S.; Rengel, Z. Aluminum-dependent dynamics of ion transport in Arabidopsis: Specificity of low pH and aluminum responses. Physiol. Plant. 2010, 139, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Rangel, A.F.; Mobin, M.; Rao, I.M.; Horst, W.J. Proton toxicity interferes with the screening of common bean (Phaseolus vulgaris L.) genotypes for aluminium resistance in nutrient solution. J. Plant Nutr. Soil Sci. 2005, 168, 607–616. [Google Scholar] [CrossRef]
- Liao, H.; Wan, H.; Shaff, J.; Wang, X.; Yan, X.; Kochian, L.V. Phosphorus and aluminum interactions in soybean in relation to aluminum tolerance. Exudation of specific organic acids from different regions of the intact root system. Plant Physiol. 2006, 141, 674–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.B.; Shen, R.F.; Zhao, X.Q.; Chen, R.F.; Dong, X.Y. Phosphorus enhances Al resistance in Al-resistant Lespedeza bicolor but not in Al-sensitive L. cuneata under relatively high Al stress. Ann. Bot. 2008, 102, 795–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, S.J.; Yang, J.L.; He, Y.F.; Yu, X.H.; Zhang, L.; You, J.F.; Shen, R.F.; Matsumoto, H. Immobilization of aluminum with phosphorus in roots is associated with high aluminum resistance in buckwheat. Plant Physiol. 2005, 138, 297–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, P.; Bai, G.; Carver, B.; Li, R.; Bernardo, A.; Baum, M. Transcriptional analysis between two wheat near-isogenic lines contrasting in aluminum tolerance under aluminum stress. Mol. Genet. Genom. 2007, 277, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.; Delhaize, E.; Jones, D. Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Biol. 2001, 52, 527–560. [Google Scholar] [CrossRef]
- Zheng, S.J.; Lin, X.; Yang, J.; Liu, Q.; Tang, C. The kinetics of aluminum adsorption and desorption by root cell walls of an aluminum resistant wheat (Triticum aestivum L.) cultivar. Plant Soil 2004, 261, 85–90. [Google Scholar] [CrossRef]
- Haridasan, M.; Paviani, T.; Schiavini, I. Localization of aluminium in the leaves of some aluminium-accumulating species. Plant Soil 1986, 94, 435–437. [Google Scholar] [CrossRef]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Ma, J.F. Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol. 2000, 41, 383–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, S.; Pinto-Carnide, O.; Martins-Lopes, P.; Matos, M.; Guedes-Pinto, H.; Santos, C. Differential aluminium changes on nutrient accumulation and root differentiation in an Al sensitive vs. tolerant wheat. Environ. Exp. Bot. 2010, 68, 91–98. [Google Scholar] [CrossRef]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Meinke, D.W.; Cherry, J.M.; Dean, C.; Rounsley, S.D.; Koornneef, M. Arabidopsis thaliana: A model plant for genome analysis. Science 1998, 282, 662–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Tandzi, N.; Ngonkeu, E.; Youmbi, E.; Nartey, E.; Yeboah, M.; Gracen, V.; Ngeve, J.; Mafouasson, H. Agronomic performance of maze hybrids under acid and control soil conditions. Int. J. Agron. Agric. Res. 2015, 6, 275–291. [Google Scholar]
- Foy, C.; Chaney, R.; White, M. The physiology of metal toxicity in plants. Annu. Rev. Plant Physiol. 1978, 29, 511–566. [Google Scholar] [CrossRef]
- Ginocchio, R.; de la Fuente, L.M.; Sánchez, P.; Bustamante, E.; Silva, Y.; Urrestarazu, P.; Rodríguez, P.H. Soil acidification as a confounding factor on metal phytotoxicity in soils spiked with copper-rich mine wastes. Environ. Toxicol. Chem. Int. J. 2009, 28, 2069–2081. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Ryan, P.R. Aluminum toxicity and tolerance in plants. Plant Physiol. 1995, 107, 315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, J.; Wen, Z.; Ru, X.; Chen, J.; Wu, H.; Wei, C. Distribution and migration of heavy metals in soil and crops affected by acid mine drainage: Public health implications in Guangdong Province, China. Ecotoxicol. Environ. Saf. 2016, 124, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Dyhr-Jensen, K.; Brix, H. Effects of pH on ammonium uptake by Typha latifolia L. Plant Cell Environ. 1996, 19, 1431–1436. [Google Scholar] [CrossRef]
- Marschner, H. Mechanisms of adaptation of plants to acid soils. Plant Soil 1991, 134, 1–20. [Google Scholar] [CrossRef]
- Felle, H.H. The apoplastic pH of the Zea mays root cortex as measured with pH-sensitive microelectrodes: Aspects of regulation. J. Exp. Bot. 1998, 49, 987–995. [Google Scholar] [CrossRef]
- Netting, A. pH, abscisic acid and the integration of metabolism in plants under stressed and non-stressed conditions. II. Modifications in modes of metabolism induced by variation in the tension on the water column and by stress. J. Exp. Bot. 2002, 53, 151–173. [Google Scholar]
- Gao, D.; Knight, M.R.; Trewavas, A.J.; Sattelmacher, B.; Plieth, C. Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol. 2004, 134, 898–908. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Rao, X.; Lu, P.; Huang, S.; Chen, X.; Xu, Z.; Xie, J. Acid-tolerant plant species screened for rehabilitating acid mine drainage sites. J. Soils Sediments 2015, 15, 1104–1112. [Google Scholar] [CrossRef]
- Fageria, N.; Morais, O.; Carvalho, M.; Colombari Filho, J. Upland rice genotype evaluations for acidity tolerance. Commun. Soil Sci. Plant Anal. 2015, 46, 1076–1096. [Google Scholar] [CrossRef]
- George, E.; Horst, W.J.; Neumann, E. Adaptation of plants to adverse chemical soil conditions. In Marschner’s Mineral Nutrition of Higher Plants; Elsevier: Amsterdam, The Netherlands, 2012; pp. 409–472. [Google Scholar]
- Gaxiola, R.A.; Fink, G.R.; Hirschi, K.D. Genetic manipulation of vacuolar proton pumps and transporters. Plant Physiol. 2002, 129, 967–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerinot, M.L. The ZIP family of metal transporters. Biochim. Biophys. Acta (BBA)-Biomembr. 2000, 1465, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Williams, L.E.; Pittman, J.K.; Hall, J. Emerging mechanisms for heavy metal transport in plants. Biochim. Biophys. Acta (BBA)-Biomembr. 2000, 1465, 104–126. [Google Scholar] [CrossRef]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.F.; Shen, R.; Nagao, S.; Tanimoto, E. Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol. 2004, 45, 583–589. [Google Scholar] [CrossRef] [Green Version]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef]
- Truernit, E.; Siemering, K.R.; Hodge, S.; Grbic, V.; Haseloff, J. A map of KNAT gene expression in the Arabidopsis root. Plant Mol. Biol. 2006, 60, 1–20. [Google Scholar] [CrossRef]
- Zhu, X.F.; Wan, J.X.; Sun, Y.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Xyloglucan endotransglucosylase-hydrolase17 interacts with xyloglucan endotransglucosylase-hydrolase31 to confer xyloglucan endotransglucosylase action and affect aluminum sensitivity in Arabidopsis. Plant Physiol. 2014, 165, 1566–1574. [Google Scholar] [CrossRef]
- Ma, J.F.; Hiradate, S.; Nomoto, K.; Iwashita, T.; Matsumoto, H. Internal detoxification mechanism of Al in hydrangea (identification of Al form in the leaves). Plant Physiol. 1997, 113, 1033–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-S.; Pi, L.-Y.; Chen, X.; Chakrabarty, P.K.; Jiang, J.; De Leon, A.L.; Liu, G.-Z.; Li, L.; Benny, U.; Oard, J. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 2006, 18, 3635–3646. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yang, C.; Li, Y.; Zheng, N.; Chen, H.; Zhao, Q.; Gao, T.; Guo, H.; Xie, Q. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 2007, 19, 1912–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adnan, A.; Mavinic, D.S.; Koch, F.A. Pilot-scale study of phosphorus recovery through struvite crystallization examining the process feasibility. J. Environ. Eng. Sci. 2003, 2, 315–324. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Z.; Gong, P.; Zhang, J.; Ziaf, K.; Li, H.; Xiao, F.; Ye, Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 2011, 33, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Combier, J.-P.; Frugier, F.; de Billy, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Vernié, T.; Ott, T.; Gamas, P.; Crespi, M. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabayek, S.; Bauer, R.; Mauerer, S.; Mizaikoff, B.; Spellerberg, B. A streptococcal NRAMP homologue is crucial for the survival of S treptococcus agalactiae under low pH conditions. Mol. Microbiol. 2016, 100, 589–606. [Google Scholar] [CrossRef] [Green Version]
- Thomine, S.; Schroeder, J.I. Plant metal transporters with homology to proteins of the NRAMP family. In The Nramp Family; Cellier, N., Gros, P., Eds.; Molecular Biology Intelligence Unit, Andes/Kluwer Series; Springer: Berlin/Heidelberg, Germany, 2004; pp. 113–121. [Google Scholar]
- Li, J.-Y.; Liu, J.; Dong, D.; Jia, X.; McCouch, S.R.; Kochian, L.V. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA 2014, 111, 6503–6508. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Piñeros, M.A.; Tian, J.; Yao, Z.; Sun, L.; Liu, J.; Shaff, J.; Coluccio, A.; Kochian, L.V.; Liao, H. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef] [Green Version]
- Rubio, F.; Arévalo, L.; Caballero, F.; Botella, M.A.; Rubio, J.S.; García-Sánchez, F.; Martínez, V. Systems involved in K+ uptake from diluted solutions in pepper plants as revealed by the use of specific inhibitors. J. Plant Physiol. 2010, 167, 1494–1499. [Google Scholar] [CrossRef]
- Lim, G.-H.; Shine, M.; de Lorenzo, L.; Yu, K.; Cui, W.; Navarre, D.; Hunt, A.G.; Lee, J.-Y.; Kachroo, A.; Kachroo, P. Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe 2016, 19, 541–549. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, D.J. Cell wall loosening by expansins. Plant Physiol. 1998, 118, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Scacchi, E.; Salinas, P.; Gujas, B.; Santuari, L.; Krogan, N.; Ragni, L.; Berleth, T.; Hardtke, C.S. Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc. Natl. Acad. Sci. USA 2010, 107, 22734–22739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gujas, B.; Alonso-Blanco, C.; Hardtke, C.S. Natural Arabidopsis brx loss-of-function alleles confer root adaptation to acidic soil. Curr. Biol. 2012, 22, 1962–1968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakazawa, M.; Yabe, N.; Ichikawa, T.; Yamamoto, Y.Y.; Yoshizumi, T.; Hasunuma, K.; Matsui, M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001, 25, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.; Biller, S.; Stanley, K.; Kajstura, T.; Prusty, R. Expression profiling of auxin-treated Arabidopsis roots: Toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 2006, 47, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.E.; Moreno-Piovano, G.; Chan, R.L. The antagonistic basic helix-loop-helix partners BEE and IBH1 contribute to control plant tolerance to abiotic stress. Plant Sci. 2018, 271, 143–150. [Google Scholar] [CrossRef]
- Li, X.; Bao, H.; Wang, Z.; Wang, M.; Fan, B.; Zhu, C.; Chen, Z. Biogenesis and function of multivesicular bodies in plant immunity. Front. Plant Sci. 2018, 9, 979. [Google Scholar] [CrossRef]
- Gleisner, M.; Kroppen, B.; Fricke, C.; Teske, N.; Kliesch, T.-T.; Janshoff, A.; Meinecke, M.; Steinem, C. Epsin N-terminal homology domain (ENTH) activity as a function of membrane tension. J. Biol. Chem. 2016, 291, 19953–19961. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, K.; Schekman, R.; Orci, L.; Heuser, J.E. Surface structure of the COPII-coated vesicle. Proc. Natl. Acad. Sci. USA 2001, 98, 13705–13709. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-K.; Shen, Y.-L.; Huang, L.-F.; Wu, S.-J.; Yeh, C.-H.; Lu, C.-A. The DEAD-box RNA helicase AtRH7/PRH75 participates in pre-rRNA processing, plant development and cold tolerance in Arabidopsis. Plant Cell Physiol. 2016, 57, 174–191. [Google Scholar] [CrossRef] [Green Version]
- Crouch, J.D.; Brosh, R.M., Jr. Mechanistic and biological considerations of oxidatively damaged DNA for helicase-dependent pathways of nucleic acid metabolism. Free Radic. Biol. Med. 2017, 107, 245–257. [Google Scholar] [CrossRef]
- Duff, S.M.; Sarath, G.; Plaxton, W.C. The role of acid phosphatases in plant phosphorus metabolism. Physiol. Plant. 1994, 90, 791–800. [Google Scholar] [CrossRef]
- Luo, H.; Laluk, K.; Lai, Z.; Veronese, P.; Song, F.; Mengiste, T. The Arabidopsis Botrytis Susceptible1 Interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 2010, 154, 1766–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, S.J.; Hocking, B.; Dayod, M.; Xu, B.; Athman, A.; Henderson, S.; Aukett, L.; Conn, V.; Shearer, M.K.; Fuentes, S. Protocol: Optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 2013, 9, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimm, D.G.; Roqueiro, D.; Salomé, P.A.; Kleebergr, S.; Greshake, B.; Zhu, W.; Liu, C.; Lippert, C.; Stegle, O.; Schölkopf, B. easyGWAS: A cloud-based platform for comparing the results of genome-wide association studies. Plant Cell 2017, 29, 5–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikka, T.; Kobayashi, Y.; Iuchi, S.; Sakurai, N.; Shibata, D.; Kobayashi, M.; Koyama, H. Natural variation of Arabidopsis thaliana reveals that aluminum resistance and proton resistance are controlled by different genetic factors. Theor. Appl. Genet. 2007, 115, 709–719. [Google Scholar] [CrossRef] [PubMed]



| Sample | pH | EC # | Al | As @ | Ba @ | Ca | Cd @ | Co @ | Cr | Cu | Fe |
| Nutrient Solution | 5.00 ± 0.03 | 1601 ± 3.61 | 0.295 ± 0.091 | <0.795 | <0.020 | 47.8 ± 1.07 | <0.051 | <0.090 | 0.158 ± 0.003 | 0.068 ± 0.006 | 3.09 ± 0.16 |
| AMD | 3.03 ± 0.01 ** | 1459 ± 2.65 * | 21.2 ± 0.453 ** | 0.032 * | 0.025 ± 0.000 * | 117 ± 4.6 ** | <0.051 | <0.090 | 0.160 ± 0.000 | 0.138 ± 0.010 ** | 53.0 ± 1.6 ** |
| Diluted AMD | 4.01 ± 0.01 ** | 1597 ± 2.31 | 0.333 ± 0.142 * | <0.795 | <0.020 | 47.1 ± 2.36 | <0.051 | <0.090 | 0.160 ± 0.000 | 0.062 ± 0.003 | 3.19 ± 0.30 * |
| Sample | K | Mg | Mn | Mo @ | Na | Ni @ | P | Pb @ | S | Se @ | Zn |
| Nutrient Solution | 242 ± 6.99 | 72.2 ± 0.32 | 0.82 ± 0.02 | <0.210 | 42.5 ± 2.61 | <0.23 | 19.8 ± 0.30 | <0.630 | 82.7 ± 1.2 | <1.13 | 1.69 ± 0.02 |
| AMD | 14.7 ± 0.72 ** | 83.1 ± 3.16 * | 9.98 ± 0.16 ** | <0.210 | 32.6 ± 2.37 ** | <0.225 | 0.283 ± 0.006 ** | <0.630 | 364 ± 17 ** | <1.125 | 0.457 ± 0.003 ** |
| Diluted AMD | 240 ± 2.7 | 71.7 ± 1.22 | 0.81 ± 0.032 | <0.210 | 43.3 ± 2.11 | <0.225 | 20.2 ± 0.94 | <0.630 | 82.9 ± 2.7 | <1.125 | 1.67 ± 0.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghimire, B.; Saminathan, T.; Bodunrin, A.; Abburi, V.L.; Kshetry, A.O.; Shinde, S.; Nimmakayala, P.; Reddy, U.K. Genome-Wide Association Study of Natural Variation in Arabidopsis Exposed to Acid Mine Drainage Toxicity and Validation of Associated Genes with Reverse Genetics. Plants 2021, 10, 191. https://doi.org/10.3390/plants10020191
Ghimire B, Saminathan T, Bodunrin A, Abburi VL, Kshetry AO, Shinde S, Nimmakayala P, Reddy UK. Genome-Wide Association Study of Natural Variation in Arabidopsis Exposed to Acid Mine Drainage Toxicity and Validation of Associated Genes with Reverse Genetics. Plants. 2021; 10(2):191. https://doi.org/10.3390/plants10020191
Chicago/Turabian StyleGhimire, Bandana, Thangasamy Saminathan, Abiodun Bodunrin, Venkata Lakshmi Abburi, Arjun Ojha Kshetry, Suhas Shinde, Padma Nimmakayala, and Umesh K. Reddy. 2021. "Genome-Wide Association Study of Natural Variation in Arabidopsis Exposed to Acid Mine Drainage Toxicity and Validation of Associated Genes with Reverse Genetics" Plants 10, no. 2: 191. https://doi.org/10.3390/plants10020191