Regulation of COX Assembly and Function by Twin CX9C Proteins—Implications for Human Disease
Abstract
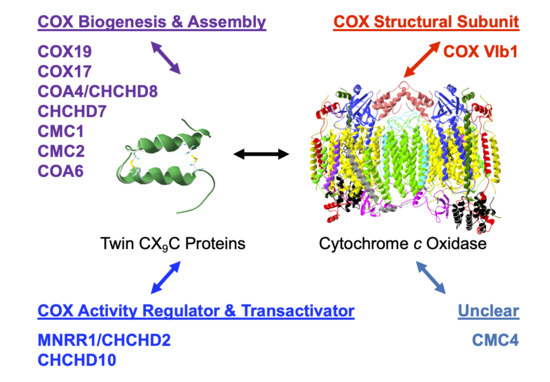
:1. Introduction
2. COX Regulation through Assembly
2.1. COX Subunit I Module Assembly
2.1.1. COX17
2.1.2. COX19
2.1.3. CHCHD7/Cox23p
2.1.4. CMC1
2.1.5. CMC2
2.1.6. COA5/Pet191p
2.2. COX Subunit II Module Assembly
COA6
2.3. Undetermined
CHCHD8/COA4
3. COX Structural Subunits
COX VIb1/Cox12p
4. COX Regulation through Direct Interaction
4.1. MNRR1 (CHCHD2)/Mix17p
4.2. CHCHD10/Mix17p
5. CX9C Proteins of Unknown Function
CMC4
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kadenbach, B.; Hüttemann, M. The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion 2015, 24, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Tsukihara, T.; Aoyama, H.; Yamashita, E.; Tomizaki, T.; Yamaguchi, H.; Shinzawa-Itoh, K.; Nakashima, R.; Yaono, R.; Yoshikawa, S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 1996, 272, 1136. [Google Scholar] [CrossRef] [PubMed]
- Osuda, Y.; Shinzawa-Itoh, K.; Tani, K.; Maeda, S.; Yoshikawa, S.; Tsukihara, T.; Gerle, C. Two-dimensional crystallization of monomeric bovine cytochrome c oxidase with bound cytochrome c in reconstituted lipid membranes. J. Electron Microsc. 2016, 65, 263–267. [Google Scholar] [CrossRef] [Green Version]
- Shinzawa-Itoh, K.; Sugimura, T.; Misaki, T.; Tadehara, Y.; Yamamoto, S.; Hanada, M.; Yano, N.; Nakagawa, T.; Uene, S.; Yamada, T.; et al. Monomeric structure of an active form of bovine cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2019, 116, 19945–19951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Gu, J.; Guo, R.; Huang, Y.; Yang, M. Structure of mammalian respiratory supercomplex I2III2IV2. Cell 2016, 167, 1598–1609.e10. [Google Scholar] [CrossRef] [Green Version]
- Balsa, E.; Marco, R.; Perales-Clemente, E.; Szklarczyk, R.; Calvo, E.; Landazuri, M.O.; Enriquez, J.A. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012, 16, 378–386. [Google Scholar] [CrossRef] [Green Version]
- Zong, S.; Wu, M.; Gu, J.; Liu, T.; Guo, R.; Yang, M. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 2018, 28, 1026–1034. [Google Scholar] [CrossRef] [Green Version]
- Kadenbach, B. Regulation of mammalian 13-subunit cytochrome c oxidase and binding of other proteins: Role of NDUFA4. Trends Endocrinol. Metab. 2017, 28, 761–770. [Google Scholar] [CrossRef]
- Cavallaro, G. Genome-wide analysis of eukaryotic twin CX9C proteins. Mol. Biosyst. 2010, 6, 2459–2470. [Google Scholar] [CrossRef]
- Glerum, D.M.; Shtanko, A.; Tzagoloff, A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 1996, 271, 14504–14509. [Google Scholar] [CrossRef] [Green Version]
- Abajian, C.; Yatsunyk La Fau-Ramirez, B.E.; Ramirez Be Fau-Rosenzweig, A.C.; Rosenzweig, A.C. Yeast Cox17 solution structure and Copper(I) binding. J. Biol. Chem. 2004, 279, 53584–53592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnesano, F.; Balatri, E.; Banci, L.; Bertini, I.; Winge, D.R. Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding. Structure 2005, 13, 713–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koehler, C.M. The small Tim proteins and the twin Cx3C motif. Trends Biochem. Sci. 2004, 29, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Terziyska, N.; Lutz, T.; Kozany, C.; Mokranjac, D.; Mesecke, N.; Neupert, W.; Herrmann, J.M.; Hell, K. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 2005, 579, 179–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chacinska, A.; Pfannschmidt, S.; Wiedemann, N.; Kozjak, V.; Sanjuan Szklarz, L.K.; Schulze-Specking, A.; Truscott, K.N.; Guiard, B.; Meisinger, C.; Pfanner, N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004, 23, 3735–3746. [Google Scholar] [CrossRef] [Green Version]
- Longen, S.; Bien, M.; Bihlmaier, K.; Kloeppel, C.; Kauff, F.; Hammermeister, M.; Westermann, B.; Herrmann, J.M.; Riemer, J. Systematic analysis of the twin CX9C protein family. J. Mol. Biol. 2009, 393, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, A.; Barros, M.H.; Valnot, I.; Rotig, A.; Rustin, P.; Tzagoloff, A. Cytochrome oxidase in health and disease. Gene 2002, 286, 53–63. [Google Scholar] [CrossRef]
- Vidoni, S.; Harbour, M.E.; Guerrero-Castillo, S.; Signes, A.; Ding, S.; Fearnley, I.M.; Taylor, R.W.; Tiranti, V.; Arnold, S.; Fernandez-Vizarra, E.; et al. MR-1S interacts with PET100 and PET117 in module-based assembly of human cytochrome c oxidase. Cell Rep. 2017, 18, 1727–1738. [Google Scholar] [CrossRef] [Green Version]
- Timon-Gomez, A.; Nyvltova, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef]
- Stiburek, L.; Vesela, K.; Hansikova, H.; Pecina, P.; Tesarova, M.; Cerna, L.; Houstek, J.; Zeman, J. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 2005, 392, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.L.; Valnot, I.; Rustin, P.; Taanman, J.W. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 2004, 279, 7462–7469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaksch, M.; Ogilvie, I.; Yao, J.; Kortenhaus, G.; Bresser, H.G.; Gerbitz, K.D.; Shoubridge, E.A. Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000, 9, 795–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaksch, M.; Paret, C.; Stucka, R.; Horn, N.; Muller-Hocker, J.; Horvath, R.; Trepesch, N.; Stecker, G.; Freisinger, P.; Thirion, C.; et al. Cytochrome c oxidase deficiency due to mutations in SCO2, encoding a mitochondrial copper-binding protein, is rescued by copper in human myoblasts. Hum. Mol. Genet. 2001, 10, 3025–3035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leary, S.C.; Kaufman, B.A.; Pellecchia, G.; Guercin, G.H.; Mattman, A.; Jaksch, M.; Shoubridge, E.A. Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 2004, 13, 1839–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, D.; Barrientos, A. Mitochondrial copper metabolism and delivery to cytochrome c oxidase. Iubmb. Life 2008, 60, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Leary, S.C.; Winge, D.R.; Cobine, P.A. Pulling the plug on cellular copper: The role of mitochondria in copper export. Biochim. Biophys Acta 2009, 1793, 146–153. [Google Scholar] [CrossRef] [Green Version]
- Valentine, J.S.; Wertz, D.L.; Lyons, T.J.; Liou, L.L.; Goto, J.J.; Gralla, E.B. The dark side of dioxygen biochemistry. Curr. Opin. Chem. Biol. 1998, 2, 253–262. [Google Scholar] [CrossRef]
- Bestwick, M.; Jeong, M.Y.; Khalimonchuk, O.; Kim, H.; Winge, D.R. Analysis of Leigh syndrome mutations in the yeast SURF1 homolog reveals a new member of the cytochrome oxidase assembly factor family. Mol. Cell. Biol. 2010, 30, 4480–4491. [Google Scholar] [CrossRef] [Green Version]
- Vogtle, F.N.; Burkhart, J.M.; Rao, S.; Gerbeth, C.; Hinrichs, J.; Martinou, J.C.; Chacinska, A.; Sickmann, A.; Zahedi, R.P.; Meisinger, C. Intermembrane space proteome of yeast mitochondria. Mol. Cell. Proteom. 2012, 11, 1840–1852. [Google Scholar] [CrossRef] [Green Version]
- Barrientos, A.; Zambrano, A.; Tzagoloff, A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004, 23, 3472–3482. [Google Scholar] [CrossRef] [Green Version]
- Fontanesi, F.; Clemente, P.; Barrientos, A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2011, 286, 555–566. [Google Scholar] [CrossRef] [Green Version]
- Mick, D.U.; Vukotic, M.; Piechura, H.; Meyer, H.E.; Warscheid, B.; Deckers, M.; Rehling, P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell. Biol. 2010, 191, 141–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemente, P.; Peralta, S.; Cruz-Bermudez, A.; Echevarria, L.; Fontanesi, F.; Barrientos, A.; Fernandez-Moreno, M.A.; Garesse, R. hCOA3 stabilizes cytochrome c oxidase 1 (COX1) and promotes cytochrome c oxidase assembly in human mitochondria. J. Biol. Chem. 2013, 288, 8321–8331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourens, M.; Barrientos, A. A CMC1-knockout reveals translation-independent control of human mitochondrial complex IV biogenesis. Embo. Rep. 2017, 18, 477–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, A.K.; Smith, D.; Gray, J.; Carr, H.S.; Liu, A.; Winge, D.R.; Hosler, J.P. Mutagenic analysis of Cox11 of Rhodobacter sphaeroides: Insights into the assembly of Cu(B) of cytochrome c oxidase. Biochemistry 2010, 49, 5651–5661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beers, J.; Glerum, D.M.; Tzagoloff, A. Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 1997, 272, 33191–33196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxfield, A.B.; Heaton, D.N.; Winge, D.R. Cox17 is functional when tethered to the mitochondrial inner membrane. J. Biol. Chem. 2004, 279, 5072–5080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glerum, D.M.; Shtanko, A.; Tzagoloff, A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 20531–20535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horng, Y.C.; Cobine, P.A.; Maxfield, A.B.; Carr, H.S.; Winge, D.R. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome c oxidase. J. Biol. Chem. 2004, 279, 35334–35340. [Google Scholar] [CrossRef] [Green Version]
- Hiser, L.; Di Valentin, M.; Hamer, A.G.; Hosler, J.P. Cox11p is required for stable formation of the Cu(B) and magnesium centers of cytochrome c oxidase. J. Biol. Chem. 2000, 275, 619–623. [Google Scholar] [CrossRef] [Green Version]
- Heaton, D.; Nittis, T.; Srinivasan, C.; Winge, D.R. Mutational analysis of the mitochondrial copper metallochaperone Cox17. J. Biol. Chem. 2000, 275, 37582–37587. [Google Scholar] [CrossRef] [Green Version]
- Heaton, D.N.; George, G.N.; Garrison, G.; Winge, D.R. The mitochondrial copper metallochaperone Cox17 exists as an oligomeric, polycopper complex. Biochemistry 2001, 40, 743–751. [Google Scholar] [CrossRef]
- Takahashi, Y.; Kako, K.; Kashiwabara, S.; Takehara, A.; Inada, Y.; Arai, H.; Nakada, K.; Kodama, H.; Hayashi, J.; Baba, T.; et al. Mammalian copper chaperone Cox17p has an essential role in activation of cytochrome c oxidase and embryonic development. Mol. Cell. Biol. 2002, 22, 7614–7621. [Google Scholar] [CrossRef] [Green Version]
- Oswald, C.; Krause-Buchholz, U.; Rödel, G. Knockdown of Human COX17 Affects assembly and supramolecular organization of cytochrome c oxidase. J. Mol. Biol. 2009, 389, 470–479. [Google Scholar] [CrossRef]
- Vanisova, M.; Burska, D.; Krizova, J.; Danhelovska, T.; Dosoudilova, Z.; Zeman, J.; Stiburek, L.; Hansikova, H. Stable COX17 downregulation leads to alterations in mitochondrial ultrastructure, decreased copper content and impaired cytochrome c oxidase biogenesis in HEK293 cells. Folia Biol. 2019, 65, 181–187. [Google Scholar]
- Palumaa, P.; Kangur, L.; Voronova, A.; Sillard, R. Metal-binding mechanism of Cox17, a copper chaperone for cytochrome c oxidase. Biochem. J. 2004, 382, 307–314. [Google Scholar] [CrossRef] [Green Version]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Janicka, A.; Martinelli, M.; Kozlowski, H.; Palumaa, P. A structural-dynamical characterization of human Cox17. J. Biol. Chem. 2008, 283, 7912–7920. [Google Scholar] [CrossRef] [Green Version]
- Cobine, P.A.; Ojeda, L.D.; Rigby, K.M.; Winge, D.R. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem. 2004, 279, 14447–14455. [Google Scholar] [CrossRef] [Green Version]
- Cobine, P.A.; Pierrel, F.; Bestwick, M.L.; Winge, D.R. Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J. Biol. Chem. 2006, 281, 36552–36559. [Google Scholar] [CrossRef] [Green Version]
- Horvath, R.; Lochmuller, H.; Stucka, R.; Yao, J.; Shoubridge, E.A.; Kim, S.H.; Gerbitz, K.D.; Jaksch, M. Characterization of human SCO1 and COX17 genes in mitochondrial cytochrome c oxidase deficiency. Biochem. Biophys. Res. Commun. 2000, 276, 530–533. [Google Scholar] [CrossRef]
- Sacconi, S.; Salviati, L.; Sue, C.M.; Shanske, S.; Davidson, M.M.; Bonilla, E.; Naini, A.B.; De Vivo, D.C.; DiMauro, S. Mutation screening in patients with isolated cytochrome c oxidase deficiency. Pediatr. Res. 2003, 53, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, M.P.; Bandeira, S.C.; Beers, J.; Tzagoloff, A. Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J. Biol. Chem. 2002, 277, 40206–40211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigby, K.; Zhang, L.; Cobine, P.A.; George, G.N.; Winge, D.R. characterization of the cytochrome c oxidase assembly factor Cox19 of Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 10233–10242. [Google Scholar] [CrossRef] [Green Version]
- Bode, M.; Woellhaf, M.W.; Bohnert, M.; van der Laan, M.; Sommer, F.; Jung, M.; Zimmermann, R.; Schroda, M.; Herrmann, J.M. Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase. Mol. Biol. Cell. 2015, 26, 2385–2401. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Horn, S.; Belkacemi, A.; Kojer, K.; Petrungaro, C.; Habich, M.; Ali, M.; Kuttner, V.; Bien, M.; Kauff, F.; et al. Protein import and oxidative folding in the mitochondrial intermembrane space of intact mammalian cells. Mol. Biol. Cell. 2013, 24, 2160–2170. [Google Scholar] [CrossRef] [PubMed]
- Tzagoloff, A.; Capitanio, N.; Nobrega, M.P.; Gatti, D. Cytochrome oxidase assembly in yeast requires the product of COX11, a homolog of the P. denitrificans protein encoded by ORF3. EMBO J. 1990, 9, 2759–2764. [Google Scholar] [CrossRef]
- Carr, H.S.; George, G.N.; Winge, D.R. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 2002, 277, 31237–31242. [Google Scholar] [CrossRef] [Green Version]
- Banci, L.; Bertini, I.; Cantini, F.; Ciofi-Baffoni, S.; Gonnelli, L.; Mangani, S. Solution structure of Cox11, a novel type of beta-immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. J. Biol. Chem. 2004, 279, 34833–34839. [Google Scholar] [CrossRef] [Green Version]
- Tay, S.K.; Nesti, C.; Mancuso, M.; Schon, E.A.; Shanske, S.; Bonilla, E.; Davidson, M.M.; Dimauro, S. Studies of COX16, COX19, and PET191 in human cytochrome c oxidase deficiency. Arch. Neurol. 2004, 61, 1935–1937. [Google Scholar] [CrossRef] [Green Version]
- Sacconi, S.; Trevisson, E.; Pistollato, F.; Baldoin, M.C.; Rezzonico, R.; Bourget, I.; Desnuelle, C.; Tenconi, R.; Basso, G.; DiMauro, S.; et al. hCOX18 and hCOX19: Two human genes involved in cytochrome c oxidase assembly. Biochem. Biophys. Res. Commun. 2005, 337, 832–839. [Google Scholar] [CrossRef]
- Leary, S.C.; Cobine, P.A.; Nishimura, T.; Verdijk, R.M.; de Krijger, R.; de Coo, R.; Tarnopolsky, M.A.; Winge, D.R.; Shoubridge, E.A. COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux. Mol. Biol. Cell. 2013, 24, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.H.; Johnson, A.; Tzagoloff, A. COX23, a homologue of COX17, is required for cytochrome oxidase assembly. J. Biol. Chem. 2004, 279, 31943–31947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dela Cruz, R.; Jeong, M.Y.; Winge, D.R. Cox1 mutation abrogates need for Cox23 in cytochrome c oxidase biogenesis. Microb. Cell. 2016, 3, 275–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Jaiswal, D.; Neri, S.; Peruzzini, R.; Winkelmann, J. Structural characterization of CHCHD5 and CHCHD7: Two atypical human twin CX9C proteins. J. Struct. Biol. 2012, 180, 190–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asp, J.; Persson, F.; Kost-Alimova, M.; Stenman, G. CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosom. Cancer 2006, 45, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.N.; Choi, P.S.; de Waal, L.; Sharifnia, T.; Imielinski, M.; Saksena, G.; Pedamallu, C.S.; Sivachenko, A.; Rosenberg, M.; Chmielecki, J.; et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS ONE 2014, 9, e87361. [Google Scholar] [CrossRef] [Green Version]
- Horn, D.; Al-Ali, H.; Barrientos, A. Cmc1p is a conserved mitochondrial twin CX9C protein involved in cytochrome c oxidase biogenesis. Mol. Cell. Biol. 2008, 28, 4354–4364. [Google Scholar] [CrossRef] [Green Version]
- Bourens, M.; Dabir, D.V.; Tienson, H.L.; Sorokina, I.; Koehler, C.M.; Barrientos, A. Role of twin Cys-Xaa9-Cys motif cysteines in mitochondrial import of the cytochrome c oxidase biogenesis factor Cmc1. J. Biol. Chem. 2012, 287, 31258–31269. [Google Scholar] [CrossRef] [Green Version]
- Mick, D.U.; Dennerlein, S.; Wiese, H.; Reinhold, R.; Pacheu-Grau, D.; Lorenzi, I.; Sasarman, F.; Weraarpachai, W.; Shoubridge, E.A.; Warscheid, B.; et al. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell 2012, 151, 1528–1541. [Google Scholar] [CrossRef] [Green Version]
- Horn, D.; Zhou, W.; Trevisson, E.; Al-Ali, H.; Harris, T.K.; Salviati, L.; Barrientos, A. The conserved mitochondrial twin CX9C protein Cmc2 is a Cmc1 homologue essential for cytochrome c oxidase biogenesis. J. Biol. Chem. 2010, 285, 15088–15099. [Google Scholar] [CrossRef] [Green Version]
- McEwen, J.E.; Hong, K.H.; Park, S.; Preciado, G.T. Sequence and chromosomal localization of two PET genes required for cytochrome c oxidase assembly in Saccharomyces cerevisiae. Curr. Genet. 1993, 23, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Baganz, F.; Hayes, A.; Farquhar, R.; Butler, P.R.; Gardner, D.C.; Oliver, S.G. Quantitative analysis of yeast gene function using competition experiments in continuous culture. Yeast 1998, 14, 1417–1427. [Google Scholar] [CrossRef]
- Khalimonchuk, O.; Rigby, K.; Bestwick, M.; Pierrel, F.; Cobine, P.A.; Winge, D.R. Pet191 is a cytochrome c oxidase assembly factor in Saccharomyces cerevisiae. Eukaryot Cell. 2008, 7, 1427–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huigsloot, M.; Nijtmans, L.G.; Szklarczyk, R.; Baars, M.J.; van den Brand, M.A.; Hendriksfranssen, M.G.; van den Heuvel, L.P.; Smeitink, J.A.; Huynen, M.A.; Rodenburg, R.J. A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am. J. Hum. Genet. 2011, 88, 488–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, Z.N.; Cobine, P.A.; Leary, S.C. The mitochondrion: A central architect of copper homeostasis. Metallomics 2017, 9, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Leary, S.C.; Sasarman, F.; Nishimura, T.; Shoubridge, E.A. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum. Mol. Genet. 2009, 18, 2230–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, A.; Trivedi, P.P.; Timbalia, S.A.; Griffin, A.T.; Rahn, J.J.; Chan, S.S.; Gohil, V.M. Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum. Mol. Genet. 2014, 23, 3596–3606. [Google Scholar] [CrossRef]
- Soma, S.; Latimer, A.J.; Chun, H.; Vicary, A.C.; Timbalia, S.A.; Boulet, A.; Rahn, J.J.; Chan, S.S.L.; Leary, S.C.; Kim, B.E.; et al. Elesclomol restores mitochondrial function in genetic models of copper deficiency. Proc. Natl. Acad. Sci. USA 2018, 115, 8161–8166. [Google Scholar] [CrossRef] [Green Version]
- Pacheu-Grau, D.; Bareth, B.; Dudek, J.; Juris, L.; Vogtle, F.N.; Wissel, M.; Leary, S.C.; Dennerlein, S.; Rehling, P.; Deckers, M. Cooperation between COA6 and SCO2 in COX2 maturation during cytochrome c oxidase assembly links two mitochondrial cardiomyopathies. Cell Metab. 2015, 21, 823–833. [Google Scholar] [CrossRef] [Green Version]
- Stroud, D.A.; Maher, M.J.; Lindau, C.; Vogtle, F.N.; Frazier, A.E.; Surgenor, E.; Mountford, H.; Singh, A.P.; Bonas, M.; Oeljeklaus, S.; et al. COA6 is a mitochondrial complex IV assembly factor critical for biogenesis of mtDNA-encoded COX2. Hum. Mol. Genet. 2015, 24, 5404–5415. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.; Pratt, A.T.; Soma, S.; Theriault, S.G.; Griffin, A.T.; Trivedi, P.P.; Gohil, V.M. Mitochondrial disease genes COA6, COX6B and SCO2 have overlapping roles in COX2 biogenesis. Hum. Mol. Genet. 2016, 25, 660–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soma, S.; Morgada, M.N.; Naik, M.T.; Boulet, A.; Roesler, A.A.; Dziuba, N.; Ghosh, A.; Yu, Q.; Lindahl, P.A.; Ames, J.B.; et al. COA6 is Structurally tuned to function as a thiol-disulfide oxidoreductase in copper delivery to mitochondrial cytochrome c oxidase. Cell Rep. 2019, 29, 4114–4126.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maghool, S.; Cooray, N.D.G.; Stroud, D.A.; Aragao, D.; Ryan, M.T.; Maher, M.J. Structural and functional characterization of the mitochondrial complex IV assembly factor Coa6. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef] [PubMed]
- Pacheu-Grau, D.; Wasilewski, M.; Oeljeklaus, S.; Gibhardt, C.S.; Aich, A.; Chudenkova, M.; Dennerlein, S.; Deckers, M.; Bogeski, I.; Warscheid, B.; et al. COA6 facilitates cytochrome c oxidase biogenesis as thiol-reductase for copper metallochaperones in mitochondria. J. Mol. Biol. 2020, 432, 2067–2079. [Google Scholar] [CrossRef] [PubMed]
- Calvo, S.E.; Compton, A.G.; Hershman, S.G.; Lim, S.C.; Lieber, D.S.; Tucker, E.J.; Laskowski, A.; Garone, C.; Liu, S.; Jaffe, D.B.; et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci. Transl. Med. 2012, 4, 118ra110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baertling, F.; M, A.M.v.d.B.; Hertecant, J.L.; Al-Shamsi, A.; L, P.v.d.H.; Distelmaier, F.; Mayatepek, E.; Smeitink, J.A.; Nijtmans, L.G.; Rodenburg, R.J. Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Hum. Mutat. 2015, 36, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Vempati, U.D.; Han, X.; Moraes, C.T. Lack of cytochrome c in mouse fibroblasts disrupts assembly/stability of respiratory complexes I and IV. J. Biol. Chem. 2009, 284, 4383–4391. [Google Scholar] [CrossRef] [Green Version]
- Barrientos, A.; Pierre, D.; Lee, J.; Tzagoloff, A. Cytochrome oxidase assembly does not require catalytically active cytochrome c. J. Biol. Chem. 2003, 278, 8881–8887. [Google Scholar] [CrossRef] [Green Version]
- Bode, M.; Longen, S.; Morgan, B.; Peleh, V.; Dick, T.P.; Bihlmaier, K.; Herrmann, J.M. Inaccurately assembled cytochrome c oxidase can lead to oxidative stress-induced growth arrest. Antioxid. Redox Signal. 2013, 18, 1597–1612. [Google Scholar] [CrossRef] [Green Version]
- LaMarche, A.E.; Abate, M.I.; Chan, S.H.; Trumpower, B.L. Isolation and characterization of COX12, the nuclear gene for a previously unrecognized subunit of Saccharomyces cerevisiae cytochrome c oxidase. J. Biol. Chem. 1992, 267, 22473–22480. [Google Scholar] [CrossRef]
- Carrero-Valenzuela, R.D.; Quan, F.; Lightowlers, R.; Kennaway, N.G.; Litt, M.; Forte, M. Human cytochrome c oxidase subunit VIb: Characterization and mapping of a multigene family. Gene 1991, 102, 229–236. [Google Scholar] [CrossRef]
- Taanman, J.W.; Schrage, C.; Reuvekamp, P.; Bijl, J.; Hartog, M.; de Vries, H.; Agsteribbe, E. Identification of three human pseudogenes for subunit VIb of cytochrome c oxidase: A molecular record of gene evolution. Gene 1991, 102, 237–244. [Google Scholar] [CrossRef]
- Hüttemann, M.; Jaradat, S.; Grossman, L.I. Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb--the counterpart to testes-specific cytochrome c? Mol. Reprod. Dev. 2003, 66, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Roberts, V.A.; Pique, M.E. Definition of the interaction domain for cytochrome c on cytochrome c oxidase. III. Prediction of the docked complex by a complete, systematic search. J. Biol. Chem. 1999, 274, 38051–38060. [Google Scholar] [CrossRef] [Green Version]
- Massa, V.; Fernandez-Vizarra, E.; Alshahwan, S.; Bakhsh, E.; Goffrini, P.; Ferrero, I.; Mereghetti, P.; D’Adamo, P.; Gasparini, P.; Zeviani, M. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am. J. Hum. Genet. 2008, 82, 1281–1289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulhag, U.N.; Soiferman, D.; Schueler-Furman, O.; Miller, C.; Shaag, A.; Elpeleg, O.; Edvardson, S.; Saada, A. Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur. J. Hum. Genet. 2015, 23, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Gabriel, K.; Milenkovic, D.; Chacinska, A.; Muller, J.; Guiard, B.; Pfanner, N.; Meisinger, C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J. Mol. Biol. 2007, 365, 612–620. [Google Scholar] [CrossRef]
- Tkach, J.M.; Yimit, A.; Lee, A.Y.; Riffle, M.; Costanzo, M.; Jaschob, D.; Hendry, J.A.; Ou, J.; Moffat, J.; Boone, C.; et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012, 14, 966–976. [Google Scholar] [CrossRef] [Green Version]
- Baughman, J.M.; Nilsson, R.; Gohil, V.M.; Arlow, D.H.; Gauhar, Z.; Mootha, V.K. A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PloS Genet. 2009, 5, e1000590. [Google Scholar] [CrossRef] [Green Version]
- Aras, S.; Pak, O.; Sommer, N.; Finley, R., Jr.; Hüttemann, M.; Weissmann, N.; Grossman, L.I. Oxygen-dependent expression of cytochrome c oxidase subunit 4–2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic. Acids. Res. 2013, 41, 2255–2266. [Google Scholar] [CrossRef] [Green Version]
- Aras, S.; Bai, M.; Lee, I.; Springett, R.; Hüttemann, M.; Grossman, L.I. MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion 2015, 20, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Aras, S.; Arrabi, H.; Purandare, N.; Hüttemann, M.; Kamholz, J.; Züchner, S.; Grossman, L.I. Abl2 kinase phosphorylates Bi-organellar regulator MNRR1 in mitochondria, stimulating respiration. Biochim. Biophys. Acta BBA Mol. Cell Res. 2017, 1864, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhou, F.; Zhang, Z.; Xing, D. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 2011, 278, 941–954. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef] [Green Version]
- MacVicar, T.; Langer, T. OPA1 processing in cell death and disease—The long and short of it. J. Cell Sci. 2016, 129, 2297–2306. [Google Scholar] [CrossRef] [Green Version]
- Floyd, B.J.; Wilkerson, E.M.; Veling, M.T.; Minogue, C.E.; Xia, C.; Beebe, E.T.; Wrobel, R.L.; Cho, H.; Kremer, L.S.; Alston, C.L.; et al. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell 2016, 63, 621–632. [Google Scholar] [CrossRef] [Green Version]
- Aras, S.; Purandare, N.; Gladyck, S.; Somayajulu-Nitu, M.; Zhang, K.; Wallace, D.C.; Grossman, L.I. Mitochondrial Nuclear Retrograde Regulator 1 (MNNR1) rescues the cellular phenotype of the MELAS mutation by inductin stress-responsive homeostatic mechanisms. Proc. Natl. Acad. Sci. USA. in Press.
- Aras, S.; Maroun, M.C.; Song, Y.; Bandyopadhyay, S.; Stark, A.; Yang, Z.Q.; Long, M.P.; Grossman, L.I.; Fernandez-Madrid, F. Mitochondrial autoimmunity and MNRR1 in breast carcinogenesis. BMC Cancer 2019, 19, 411. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Vellanki, R.N.; Coyaud, E.; Ignatchenko, V.; Li, L.; Krieger, J.R.; Taylor, P.; Tong, J.; Pham, N.A.; Liu, G.; et al. CHCHD2 Is coamplified with EGFR in NSCLC and regulates mitochondrial function and cell migration. Mol. Cancer Res. 2015, 13, 1119–1129. [Google Scholar] [CrossRef] [Green Version]
- Song, R.; Yang, B.; Gao, X.; Zhang, J.; Sun, L.; Wang, P.; Meng, Y.; Wang, Q.; Liu, S.; Cheng, J. Cyclic adenosine monophosphate response element-binding protein transcriptionally regulates CHCHD2 associated with the molecular pathogenesis of hepatocellular carcinoma. Mol. Med.Rep. 2015, 11, 4053–4062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funayama, M.; Ohe, K.; Amo, T.; Furuya, N.; Yamaguchi, J.; Saiki, S.; Li, Y.; Ogaki, K.; Ando, M.; Yoshino, H.; et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson’s disease: A genome-wide linkage and sequencing study. Lancet Neurol. 2015, 14, 274–282. [Google Scholar] [CrossRef]
- Feyeux, M.; Bourgois-Rocha, F.; Redfern, A.; Giles, P.; Lefort, N.; Aubert, S.; Bonnefond, C.; Bugi, A.; Ruiz, M.; Deglon, N.; et al. Early transcriptional changes linked to naturally occurring Huntington’s disease mutations in neural derivatives of human embryonic stem cells. Hum. Mol. Genet. 2012, 21, 3883–3895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimojima, K.; Okumura, A.; Hayashi, M.; Kondo, T.; Inoue, H.; Yamamoto, T. CHCHD2 is down-regulated in neuronal cells differentiated from iPS cells derived from patients with lissencephaly. Genomics 2015, 106, 196–203. [Google Scholar] [CrossRef]
- Seo, M.; Lee, W.H.; Suk, K. Identification of novel cell migration-promoting genes by a functional genetic screen. FASEB J. 2010, 24, 464–478. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Wu, B.P.; Nguyen, D.; Liu, Y.T.; Marani, M.; Hench, J.; Benit, P.; Kozjak-Pavlovic, V.; Rustin, P.; Frank, S.; et al. CHCHD2 accumulates in distressed mitochondria and facilitates oligomerization of CHCHD10. Hum. Mol. Genet. 2018, 27, 3881–3900. [Google Scholar] [CrossRef]
- Hüttemann, M.; Lee, I.; Liu, J.; Grossman, L.I. Transcription of mammalian cytochrome c oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J. 2007, 274, 5737–5748. [Google Scholar] [CrossRef]
- Hüttemann, M.; Kadenbach, B.; Grossman, L.I. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene 2001, 267, 111–123. [Google Scholar] [CrossRef]
- Moreno-Dominguez, A.; Ortega-Saenz, P.; Gao, L.; Colinas, O.; Garcia-Flores, P.; Bonilla-Henao, V.; Aragones, J.; Hüttemann, M.; Grossman, L.I.; Weissmann, N.; et al. Acute O2 sensing through HIF2alpha-dependent expression of atypical cytochrome oxidase subunits in arterial chemoreceptors. Sci. Signal. 2020, 13. [Google Scholar] [CrossRef] [Green Version]
- Hüttemann, M.; Lee, I.; Gao, X.; Pecina, P.; Pecinova, A.; Liu, J.; Aras, S.; Sommer, N.; Sanderson, T.H.; Tost, M.; et al. Cytochrome c oxidase subunit 4 isoform 2-knockout mice show reduced enzyme activity, airway hyporeactivity, and lung pathology. FASEB J. 2012, 26, 3916–3930. [Google Scholar] [CrossRef] [Green Version]
- Martherus, R.S.; Sluiter, W.; Timmer, E.D.; VanHerle, S.J.; Smeets, H.J.; Ayoubi, T.A. Functional annotation of heart enriched mitochondrial genes GBAS and CHCHD10 through guilt by association. Biochem. Biophys. Res. Commun. 2010, 402, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Ajroud-Driss, S.; Fecto, F.; Ajroud, K.; Lalani, I.; Calvo, S.E.; Mootha, V.K.; Deng, H.X.; Siddique, N.; Tahmoush, A.J.; Heiman-Patterson, T.D.; et al. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics 2015, 16, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Purandare, N.; Somayajulu, M.; Hüttemann, M.; Grossman, L.I.; Aras, S. The cellular stress proteins CHCHD10 and MNRR1 (CHCHD2): Partners in mitochondrial and nuclear function and dysfunction. J. Biol. Chem. 2018, 293, 6517–6529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannwarth, S.; Ait-El-Mkadem, S.; Chaussenot, A.; Genin, E.C.; Lacas-Gervais, S.; Fragaki, K.; Berg-Alonso, L.; Kageyama, Y.; Serre, V.; Moore, D.G.; et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 2014, 137, 2329–2345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penttila, S.; Jokela, M.; Bouquin, H.; Saukkonen, A.M.; Toivanen, J.; Udd, B. Late onset spinal motor neuronopathy is caused by mutation in CHCHD10. Ann. Neurol. 2015, 77, 163–172. [Google Scholar] [CrossRef]
- Chaussenot, A.; Le Ber, I.; Ait-El-Mkadem, S.; Camuzat, A.; de Septenville, A.; Bannwarth, S.; Genin, E.C.; Serre, V.; Auge, G.; French research network on, F.T.D.; et al. Screening of CHCHD10 in a French cohort confirms the involvement of this gene in frontotemporal dementia with amyotrophic lateral sclerosis patients. Neurobiol. Aging 2014, 35, 2884.e1–2884.e4. [Google Scholar] [CrossRef]
- Muller, K.; Andersen, P.M.; Hubers, A.; Marroquin, N.; Volk, A.E.; Danzer, K.M.; Meitinger, T.; Ludolph, A.C.; Strom, T.M.; Weishaupt, J.H. Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease. Brain 2014, 137, e309. [Google Scholar] [CrossRef] [Green Version]
- Auranen, M.; Ylikallio, E.; Shcherbii, M.; Paetau, A.; Kiuru-Enari, S.; Toppila, J.P.; Tyynismaa, H. CHCHD10 variant p.(Gly66Val) causes axonal Charcot-Marie-Tooth disease. Neurol. Genet. 2015, 1, e1. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Salokas, K.; Tamene, F.; Jiu, Y.; Weldatsadik, R.G.; Ohman, T.; Varjosalo, M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018, 9, 1188. [Google Scholar] [CrossRef] [Green Version]
- Madani, A.; Choukroun, V.; Soulier, J.; Cacheux, V.; Claisse, J.F.; Valensi, F.; Daliphard, S.; Cazin, B.; Levy, V.; Leblond, V.; et al. Expression of p13MTCP1 is restricted to mature T-cell proliferations with t(X;14) translocations. Blood 1996, 87, 1923–1927. [Google Scholar] [CrossRef] [Green Version]
- Madani, A.; Soulier, J.; Schmid, M.; Plichtova, R.; Lerme, F.; Gateau-Roesch, O.; Garnier, J.P.; Pla, M.; Sigaux, F.; Stern, M.H. The 8 kD product of the putative oncogene MTCP-1 is a mitochondrial protein. Oncogene 1995, 10, 2259–2262. [Google Scholar] [PubMed]
- Soulier, J.; Madani, A.; Cacheux, V.; Rosenzwajg, M.; Sigaux, F.; Stern, M.H. The MTCP-1/c6.1B gene encodes for a cytoplasmic 8 kD protein overexpressed in T cell leukemia bearing a t(X;14) translocation. Oncogene 1994, 9, 3565–3570. [Google Scholar] [PubMed]
- Stern, M.H.; Soulier, J.; Rosenzwajg, M.; Nakahara, K.; Canki-Klain, N.; Aurias, A.; Sigaux, F.; Kirsch, I.R. MTCP-1: A novel gene on the human chromosome Xq28 translocated to the T cell receptor alpha/delta locus in mature T cell proliferations. Oncogene 1993, 8, 2475–2483. [Google Scholar] [PubMed]
- Fisch, P.; Forster, A.; Sherrington, P.D.; Dyer, M.J.; Rabbitts, T.H. The chromosomal translocation t(X;14)(q28;q11) in T-cell pro-lymphocytic leukaemia breaks within one gene and activates another. Oncogene 1993, 8, 3271–3276. [Google Scholar] [PubMed]
- Schoenfeld, R.A.; Napoli, E.; Wong, A.; Zhan, S.; Reutenauer, L.; Morin, D.; Buckpitt, A.R.; Taroni, F.; Lonnerdal, B.; Ristow, M.; et al. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum. Mol. Genet. 2005, 14, 3787–3799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Vizarra, E.; Tiranti, V.; Zeviani, M. Assembly of the oxidative phosphorylation system in humans: What we have learned by studying its defects. Biochim. Biophys. Acta. 2009, 1793, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Pecina, P.; Houstkova, H.; Hansikova, H.; Zeman, J.; Houstek, J. Genetic defects of cytochrome c oxidase assembly. Physiol. Res. 2004, 53 (Suppl 1), S213–S223. [Google Scholar]
- Barrientos, A.; Gouget, K.; Horn, D.; Soto, I.C.; Fontanesi, F. Suppression mechanisms of COX assembly defects in yeast and human: Insights into the COX assembly process. Biochim. Biophys. Acta. 2009, 1793, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Soto, I.C.; Fontanesi, F.; Liu, J.; Barrientos, A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta. 2012, 1817, 883–897. [Google Scholar] [CrossRef] [Green Version]
- Valnot, I.; Osmond, S.; Gigarel, N.; Mehaye, B.; Amiel, J.; Cormier-Daire, V.; Munnich, A.; Bonnefont, J.P.; Rustin, P.; Rotig, A. Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet. 2000, 67, 1104–1109. [Google Scholar] [CrossRef] [Green Version]
- Papadopoulou, L.C.; Sue, C.M.; Davidson, M.M.; Tanji, K.; Nishino, I.; Sadlock, J.E.; Krishna, S.; Walker, W.; Selby, J.; Glerum, D.M.; et al. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999, 23, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Sue, C.M.; Karadimas, C.; Checcarelli, N.; Tanji, K.; Papadopoulou, L.C.; Pallotti, F.; Guo, F.L.; Shanske, S.; Hirano, M.; De Vivo, D.C.; et al. Differential features of patients with mutations in two COX assembly genes, SURF-1 and SCO2. Ann. Neurol. 2000, 47, 589–595. [Google Scholar] [CrossRef]
- Salviati, L.; Sacconi, S.; Rasalan, M.M.; Kronn, D.F.; Braun, A.; Canoll, P.; Davidson, M.; Shanske, S.; Bonilla, E.; Hays, A.P.; et al. Cytochrome c oxidase deficiency due to a novel SCO2 mutation mimics Werdnig-Hoffmann disease. Arch. Neurol. 2002, 59, 862–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valnot, I.; von Kleist-Retzow, J.C.; Barrientos, A.; Gorbatyuk, M.; Taanman, J.W.; Mehaye, B.; Rustin, P.; Tzagoloff, A.; Munnich, A.; Rotig, A. A mutation in the human heme A:farnesyltransferase gene (COX10 ) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000, 9, 1245–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonicka, H.; Leary, S.C.; Guercin, G.H.; Agar, J.N.; Horvath, R.; Kennaway, N.G.; Harding, C.O.; Jaksch, M.; Shoubridge, E.A. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 2003, 12, 2693–2702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonicka, H.; Mattman, A.; Carlson, C.G.; Glerum, D.M.; Hoffbuhr, K.C.; Leary, S.C.; Kennaway, N.G.; Shoubridge, E.A. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2003, 72, 101–114. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Yao, J.; Johns, T.; Fu, K.; De Bie, I.; Macmillan, C.; Cuthbert, A.P.; Newbold, R.F.; Wang, J.; Chevrette, M.; et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998, 20, 337–343. [Google Scholar] [CrossRef]
- Tiranti, V.; Hoertnagel, K.; Carrozzo, R.; Galimberti, C.; Munaro, M.; Granatiero, M.; Zelante, L.; Gasparini, P.; Marzella, R.; Rocchi, M.; et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 1998, 63, 1609–1621. [Google Scholar] [CrossRef] [Green Version]
- Naoe, M.; Ohwa, Y.; Ishikawa, D.; Ohshima, C.; Nishikawa, S.; Yamamoto, H.; Endo, T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 2004, 279, 47815–47821. [Google Scholar] [CrossRef] [Green Version]
- Mesecke, N.; Terziyska, N.; Kozany, C.; Baumann, F.; Neupert, W.; Hell, K.; Herrmann, J.M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 2005, 121, 1059–1069. [Google Scholar] [CrossRef] [Green Version]
- Allen, S.; Balabanidou, V.; Sideris, D.P.; Lisowsky, T.; Tokatlidis, K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 2005, 353, 937–944. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic. Acids. Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bak, D.W.; Weerapana, E. Cysteine-mediated redox signalling in the mitochondria. Mol. Biosyst. 2015, 11, 678–697. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jedrychowski, M.P.; Schweppe, D.K.; Huttlin, E.L.; Yu, Q.; Heppner, D.E.; Li, J.; Long, J.; Mills, E.L.; Szpyt, J.; et al. A quantitative tissue-specific landscape of protein redox regulation during aging. Cell 2020, 180, 968–983. [Google Scholar] [CrossRef] [PubMed]













| Types of Regulation of COX |
|---|
| Expression of tissue-, developmental-, and/or species-specific isoforms of subunits |
| Interaction with small molecules |
| Reversible phosphorylation of subunits |
| Protein–protein interactions |
| Supercomplex formation |
| Human Protein | Yeast Protein | Function |
|---|---|---|
| CX9C Proteins | ||
| COX17 | Cox17p | COX copper chaperone |
| COX19 | Cox19p | COX assembly |
| CMC1 | Cmc1p | COX assembly |
| CMC2 | Cmc2p | COX assembly |
| COA5 | Pet191p | COX assembly |
| COA6 | Coa6p | COX assembly |
| CHCHD7 | Cox23p | COX assembly |
| CHCHD8 | Coa4p | COX/complex III assembly/function |
| MNRR1/CHCHD2 | Mix17p | Activity regulation |
| CHCHD10 | Mix17p | Activity regulation |
| CMC4 | Cmc4p | Unknown |
| COX VIb1 | Cox12p | Subunit |
| Cytochrome c Oxidase | ||
| COX I | Cox1p | Subunit |
| COX II | Cox2p | |
| COX III | Cox3p | |
| COX IV | Cox4p | |
| COX Va | Cox5Ap | |
| COX Vb | Cox5Bp | |
| COX VI | Cox6p | |
| COX VII | Cox7p | |
| COX VIII | COX8p | |
| COX IX | Cox9p | |
| COX XIII | Cox13p | |
| Protein | Disease Association | Reference |
|---|---|---|
| COX17 | COX17-null embryonic lethal in mice | [43] |
| COX19 | None | |
| CHCHD7 | Salivary gland pleiomorphic adenomas (benign) | [65] |
| Lung adenocarcinoma | [66] | |
| Acute myeloid leukemia | [66] | |
| CMC1 | None | |
| CMC2 | None | |
| COA5 | Hypertrophic cardiomyopathy | [67] |
| COA6 | Hypertrophic cardiomyopathy | [68,69] |
| CHCHD8 | None | |
| MNRR1/CHCHD2 | Parkinson’s disease | [70] |
| Huntington’s disease | [71] | |
| Lissencephaly | [72] | |
| Breast cancer | [73] | |
| EGFR-positive non-small cell lung cancer | [74] | |
| Hepatitis B or C virus associated hepatocellular carcinoma | [75] | |
| CHCHD10 | Spinal muscular atrophy, Jokela type | [76] |
| ALS and/or front temporal dementia | [77,78] | |
| Autosomal dominant mitochondrial myopathy | [79] | |
| Lower motor neuron syndrome | [80] | |
| Charcot-Marie-Tooth disease type 2 | [81] | |
| CMC4 | T cell leukemia with translocation (X:14) | [82,83,84,85,86] |
| Frataxin mRNA deficiency | [87] | |
| COXVIb1 | COX deficiency/mitochondrial encephalomyopathy | [88,89] |
| ID/Sequence Window Parameters | Gene | ORE |
|---|---|---|
| Published ORE | COXIVI2 | GGACGT TCCCA CGCTGGGGCGG |
| NC_000015.10:74937913-74938533 | COXVa | TCGGCC TCCCA AAAGTGCTGGG |
| CTGTAA TCCCA GCACTTTTGGG | ||
| NC_000002.12:97645525-97646190 | COXVb | TCCGCC TCCCA CACACAGTACA |
| AGACTT TCCCA CCTCCCAGCGT | ||
| CCCACC TCCCA GCGTTATTAAA | ||
| GCCCAC TCCCA AGACTGTGGCG | ||
| GCTTCC TCCCA GCCCTCGAGCC | ||
| NC_000012.12:120437600-120438231 | COXVIa | TTCTGA TCCCA GACCGCCCTAT |
| CCGCCC TCCCA CTGGCATTAAC | ||
| NC_000029.10:3560717-35651348 | COXVIb1 | GATGAA TCCCA GAAACCTCTCG |
| NC_000008.11:99891909-99892550 | COXVIc | CCGGAA TCCCA GCACTTTGGGA |
| TCGTCC TCCCA AAGTGCTGGGA | ||
| NC_000019.10:36152390-36153000 | COXVIIa | TTTTAC TCCCA AAGACTTTAGC |
| GCTTGG TCCCA CCGGAGCCCAA | ||
| NC_000023.11:77899014-77899593 | COXVIIb | TACATT TCCCA AAAGGCCTTTA |
| NC_000005.10:86617391-86618131 | COXVIIc | TTCGAT TCCCA TTATCTTTAGT |
| TACATT TCCCA CAATCCTTCGG | ||
| TACAAT TCCCA CAATCCAGGGC | ||
| CCCATT TCCCA TCTTTCTTTTC | ||
| TAACCT TCCCA GGGCCCTCCTC | ||
| NC_000011.10:63974100-63974795 | COXVIIIa | CCGGCC TCCCA AAGTGCTGGGA |
| CTCTAA TCCCA GCACTTTGGGA | ||
| ACGGCC TCCCA AAAGTTCTGAG | ||
| CACACC TCCCA CAGCCCTTGGC | ||
| CCATGA TCCCA AGCTTCCCCTC | ||
| NC_000017.11:14068795-14069649 | COX10 | CTGGGC TCCCA TTTGATAGAGA |
| TCGACC TCCCA GGCTCAAGCGA | ||
| TCGAGT TCCCA GGATCAAGCGA | ||
| CTGCAA TCCCA GTGCTTAGGGA | ||
| TGCAGC TCCCA GAAGTTGACAG | ||
| NC_000017.11:54968279-54969164 | COX11 | TGTTAC TCCCA CTTCGCTTCTC |
| NC_000012.12:50119498-50120218 | COX14 | TCAGCC TCCCA AGTAGCTGACG |
| CCACCC TCCCA CCTCAGCCTCC | ||
| NC_000010.11:99731958-99732640 | COX15 | None |
| NC_000003.12:119677203-119677899 | COX17 | None |
| NC_000004.12:73069629-73030150 | COX18 | CGCCTT TCCCA CGCGGCACTGG |
| TGACCT TCCCA GTCAAGCCGGT | ||
| NC_000001.11:244834975-244835753 | COX20 | CGGGGC TCCCA CGCCCACCCCA |
| NC_000007.14:43647530-4348140 | COA1 | TCCTCA TCCCA AGGGTGGTGCA |
| TAGGAT TCCCA GTTTTGTGCCA | ||
| CCTTGC TCCCA GAGGCATTGAC | ||
| NC_000011.10:73873321-73874201 | COA4 | TCAGTC TCCCA AAGTGCTGGGA |
| CTATAA TCCCA GCACTTTGGGA | ||
| TCAGCC TCCCA TAATGCTAGGA | ||
| CTGTAA TCCCA GCATTTTGGGT | ||
| CTGTAG TCCCA GCTACTCTGGC | ||
| NC_000001.11:234372861-234372961 | COA6 | TTATTT TCCCA ACTCCCCTGCC |
| GTGACG TCCCA GAACCCATGGC | ||
| CTGTAG TCCCA GCTACCTGGGA | ||
| CCTGCC TCCCA GGTAGCTGGGA | ||
| AGATCC TCCCA CCTCAGCCTGC | ||
| NC_000003.12:28241299-28241909 | CMC1 | GCACAG TCCCA AAGTCCTGCAG |
| GCACAG TCCCA AAGTCCTGCAG | ||
| CTTCCT TCCCA GGCTCCTACTT | ||
| CACCTT TCCCA GGTCGGCATCC | ||
| NC_000016.10:80997314-80997940 | CMC2 | None |
| NC_000023.11:155063965-155064531 | CMC4 | TATTAT TCCCA AGACTTTTTTT |
| TATGAT TCCCA TTTTATATATG | ||
| TACTCT TCCCA AACATAGGATG | ||
| NC_000009.12:133356399-133357002 | SURF1 | CCCTCA TCCCA ACTGCGCCCTT |
| CACCCG TCCCA GCCCCGCCGCC | ||
| NC_000003.12:42804144-42804699 | HIGD1A | CAGCCG TCCCA GCCAATCAGAG |
| ACGGAC TCCCA GCCCCCACCCC | ||
| NC_000007.14:10940000-10940611 | NDUFA4 | CTCGGC TCCCA GAGGCGCCGCG |
| CTGCCC TCCCA GCCAAGGGTCC | ||
| NC_000007.14:56106405-56106909 | CHCHD2/MNRR1 | CCCGCC TCCCA TCTTCCGGTCT |
| TGGTTG TCCCA CGTCCGGAGGC | ||
| NC_000008.11:56212000-56212924 | CHCHD7 | CTGATT TCCCA TCCAGTGGTTC |
| NC_000022.11:23767863-237687376 | CHCHD10 | TCCCCT TCCCA GCTGGCCCCAT |
| GCAGGG TCCCA TTTCCAGCCGA | ||
| CCCGGG TCCCA CCGCCGCCACC | ||
| NC_000020.11:18137355-18138051 | PET117 | CGGGGT TCCCA TCCGTGGAGTT |
| CCCCTC TCCCA CAGACGTCCCT | ||
| NC_000017.11:10697489-10697998 | SCO1 | None |
| NC_000022:11:50524401-50524931 | SCO2 | CCACAC TCCCA GCAGAAAACCT |
| TGGTGA TCCCA GGTAGAGGACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gladyck, S.; Aras, S.; Hüttemann, M.; Grossman, L.I. Regulation of COX Assembly and Function by Twin CX9C Proteins—Implications for Human Disease. Cells 2021, 10, 197. https://doi.org/10.3390/cells10020197
Gladyck S, Aras S, Hüttemann M, Grossman LI. Regulation of COX Assembly and Function by Twin CX9C Proteins—Implications for Human Disease. Cells. 2021; 10(2):197. https://doi.org/10.3390/cells10020197
Chicago/Turabian StyleGladyck, Stephanie, Siddhesh Aras, Maik Hüttemann, and Lawrence I. Grossman. 2021. "Regulation of COX Assembly and Function by Twin CX9C Proteins—Implications for Human Disease" Cells 10, no. 2: 197. https://doi.org/10.3390/cells10020197