Abstract
Purpose
Cholinergic signals can be important modulators of cellular signaling in cancer. We recently have shown that knockdown of nicotinic acetylcholine receptor subunit alpha 5, CHRNA5, diminishes the proliferative potential of breast cancer cells. However, modulation of CHRNA5 expression in the context of estrogen signaling and its prognostic implications in breast cancer remained unexplored.
Methods
Meta-analyses of large breast cancer microarray cohorts were used to evaluate the association of CHRNA5 expression with estrogen (E2) treatment, estrogen receptor (ER) status and patient prognosis. The results were validated through RT-qPCR analyses of multiple E2 treated cell lines, CHRNA5 depleted MCF7 cells and across a breast cancer patient cDNA panel. We also calculated a predicted secondary (PS) score representing direct/indirect induction of gene expression by E2 based on a public dataset (GSE8597). Co-expression analysis was performed using a weighted gene co-expression network analysis (WGCNA) pipeline. Multiple other publicly available datasets such as CCLE, COSMIC and TCGA were also analyzed.
Results
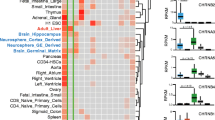
Herein we found that CHRNA5 expression was induced by E2 in a dose- and time-dependent manner in breast cancer cell lines. ER− breast tumors exhibited higher CHRNA5 expression levels than ER+ tumors. Independent meta-analysis for survival outcome revealed that higher CHRNA5 expression was associated with a worse prognosis in untreated breast cancer patients. Furthermore, CHRNA5 and its co-expressed gene network emerged as secondarily induced targets of E2 stimulation. These targets were largely downregulated by exposure to CHRNA5 siRNA in MCF7 cells while the response of primary ESR1 targets was dependent on the direction of the PS-score. Moreover, primary and secondary target genes were uncoupled and clustered distinctly based on multiple public datasets.
Conclusion
Our findings strongly associate increased expression of CHRNA5 and its co-expression network with secondary E2 signaling and a worse prognosis in breast cancer.










Similar content being viewed by others
References
T. Truong, R.J. Hung, C.I. Amos, X. Wu, H. Bickeboller, A. Rosenberger, W. Sauter, T. Illig, H.E. Wichmann, A. Risch, H. Dienemann, R. Kaaks, P. Yang, R. Jiang, J.K. Wiencke, M. Wrensch, H. Hansen, K.T. Kelsey, K. Matsuo, K. Tajima, A.G. Schwartz, A. Wenzlaff, A. Seow, C. Ying, A. Staratschek-Jox, P. Nurnberg, E. Stoelben, J. Wolf, P. Lazarus, J.E. Muscat, C.J. Gallagher, S. Zienolddiny, A. Haugen, H.F. van der Heijden, L.A. Kiemeney, D. Isla, J.I. Mayordomo, T. Rafnar, K. Stefansson, Z.F. Zhang, S.C. Chang, J.H. Kim, Y.C. Hong, E.J. Duell, A.S. Andrew, F. Lejbkowicz, G. Rennert, H. Muller, H. Brenner, L. Le Marchand, S. Benhamou, C. Bouchardy, M.D. Teare, X. Xue, J. McLaughlin, G. Liu, J.D. McKay, P. Brennan, M.R. Spitz, Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst 102, 959–971 (2010). https://doi.org/10.1093/jnci/djq178
F.S. Falvella, A. Galvan, E. Frullanti, M. Spinola, E. Calabro, A. Carbone, M. Incarbone, L. Santambrogio, U. Pastorino, T.A. Dragani, Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin Cancer Res 15, 1837–1842 (2009). https://doi.org/10.1158/1078-0432.CCR-08-2107
S.S. Yoo, S.M. Lee, S.K. Do, W.K. Lee, D.S. Kim, J.Y. Park, Unmethylation of the CHRNB4 gene is an unfavorable prognostic factor in non-small cell lung cancer. Lung Cancer 86, 85–90 (2014). https://doi.org/10.1016/j.lungcan.2014.08.002
F.S. Falvella, T. Alberio, S. Noci, L. Santambrogio, M. Nosotti, M. Incarbone, U. Pastorino, M. Fasano, T.A. Dragani, Multiple isoforms and differential allelic expression of CHRNA5 in lung tissue and lung adenocarcinoma. Carcinogenesis 34, 1281–1285 (2013). https://doi.org/10.1093/carcin/bgt062
C.C. Warzecha, S. Shen, Y. Xing, R.P. Carstens, The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol 6, 546–562 (2009)
Q. Xu, T. Hamada, R. Kiyama, Y. Sakuma, Y. Wada-Kiyama, Site-specific regulation of gene expression by estrogen in the hypothalamus of adult female rats. Neurosci Lett 436, 35–39 (2008). https://doi.org/10.1016/j.neulet.2008.02.054
C.H. Lee, Y.C. Chang, C.S. Chen, S.H. Tu, Y.J. Wang, L.C. Chen, Y.J. Chang, P.L. Wei, H.W. Chang, C.H. Chang, C.S. Huang, C.H. Wu, Y.S. Ho, Crosstalk between nicotine and estrogen-induced estrogen receptor activation induces alpha9-nicotinic acetylcholine receptor expression in human breast cancer cells. Breast Cancer Res Treat 129, 331–345 (2011)
C.H. Lee, C.S. Huang, C.S. Chen, S.H. Tu, Y.J. Wang, Y.J. Chang, K.W. Tam, P.L. Wei, T.C. Cheng, J.S. Chu, L.C. Chen, C.H. Wu, Y.S. Ho, Overexpression and activation of the alpha9-nicotinic receptor during tumorigenesis in human breast epithelial cells. J Natl Cancer Inst 102, 1322–1335 (2010). https://doi.org/10.1093/jnci/djq300
S. Cingir Koker, E. Jahja, H. Shehwana, A.G. Keskus, O. Konu, Cholinergic receptor nicotinic alpha 5 (CHRNA5) RNAi is associated with cell cycle inhibition, apoptosis. DNA damage response and drug sensitivity in breast cancer PLoS One 13, e0208982 (2018). https://doi.org/10.1371/journal.pone.0208982
M.R. Meyer, E. Haas, E.R. Prossnitz, M. Barton, Non-genomic regulation of vascular cell function and growth by estrogen. Mol Cell Endocrinol 308, 9–16 (2009). https://doi.org/10.1016/j.mce.2009.03.009
J. Frasor, J.M. Danes, B. Komm, K.C. Chang, C.R. Lyttle, B.S. Katzenellenbogen, Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: Insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144, 4562–4574 (2003). https://doi.org/10.1210/en.2003-0567
V. Bourdeau, J. Deschenes, D. Laperriere, M. Aid, J.H. White, S. Mader, Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res 36, 76–93 (2008). https://doi.org/10.1093/nar/gkm945
V. Jagannathan, M. Robinson-Rechavi, Meta-analysis of estrogen response in MCF-7 distinguishes early target genes involved in signaling and cell proliferation from later target genes involved in cell cycle and DNA repair. BMC Syst Biol 5, 138 (2011). https://doi.org/10.1186/1752-0509-5-138
B.K. Banin Hirata, J.M. Oda, R. Losi Guembarovski, C.B. Ariza, C.E. de Oliveira, M.A. Watanabe, Molecular markers for breast cancer: Prediction on tumor behavior. Dis Markers 2014, 513158–513112 (2014). https://doi.org/10.1155/2014/513158
E.R. Prossnitz, M. Maggiolini, Mechanisms of estrogen signaling and gene expression via GPR30. Mol Cell Endocrinol 308, 32–38 (2009). https://doi.org/10.1016/j.mce.2009.03.026
B. Haibe-Kains, C. Desmedt, F. Piette, M. Buyse, F. Cardoso, L. Van't Veer, M. Piccart, G. Bontempi, C. Sotiriou, Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics 9, 394 (2008). https://doi.org/10.1186/1471-2164-9-394
E.K. Millar, P.H. Graham, S.A. O'Toole, C.M. McNeil, L. Browne, A.L. Morey, S. Eggleton, J. Beretov, C. Theocharous, A. Capp, E. Nasser, J.H. Kearsley, G. Delaney, G. Papadatos, C. Fox, R.L. Sutherland, Prediction of local recurrence, distant metastases, and death after breast-conserving therapy in early-stage invasive breast cancer using a five-biomarker panel. J Clin Oncol 27, 4701–4708 (2009). https://doi.org/10.1200/JCO.2008.21.7075
M.L. Salmans, F. Zhao, B. Andersen, The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: A potential drug target and biomarker. Breast Cancer Res 15, 204 (2013). https://doi.org/10.1186/bcr3408
D.S. Oh, M.A. Troester, J. Usary, Z. Hu, X. He, C. Fan, J. Wu, L.A. Carey, C.M. Perou, Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol 24, 1656–1664 (2006). https://doi.org/10.1200/JCO.2005.03.2755
R. Kumar, A. Sharma, R.K. Tiwari, Application of microarray in breast cancer: An overview. J Pharm Bioallied Sci 4, 21–26 (2012). https://doi.org/10.4103/0975-7406.92726
B.J. Trock, F. Leonessa, R. Clarke, Multidrug resistance in breast cancer: A meta-analysis of MDR1/gp170 expression and its possible functional significance. J Natl Cancer Inst 89, 917–931 (1997)
Y. Pawitan, J. Bjohle, L. Amler, A.L. Borg, S. Egyhazi, P. Hall, X. Han, L. Holmberg, F. Huang, S. Klaar, E.T. Liu, L. Miller, H. Nordgren, A. Ploner, K. Sandelin, P.M. Shaw, J. Smeds, L. Skoog, S. Wedren, J. Bergh, Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res 7, R953–R964 (2005). https://doi.org/10.1186/bcr1325
T. Barrett, D.B. Troup, S.E. Wilhite, P. Ledoux, C. Evangelista, I.F. Kim, M. Tomashevsky, K.A. Marshall, K.H. Phillippy, P.M. Sherman, R.N. Muertter, M. Holko, O. Ayanbule, A. Yefanov, A. Soboleva, NCBI GEO: Archive for functional genomics data sets--10 years on. Nucleic Acids Res 39, D1005–D1010 (2011). https://doi.org/10.1093/nar/gkq1184
R. Edgar, M. Domrachev, A.E. Lash, Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210 (2002)
B. Gyorffy, A. Lanczky, A.C. Eklund, C. Denkert, J. Budczies, Q. Li, Z. Szallasi, An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123, 725–731 (2010). https://doi.org/10.1007/s10549-009-0674-9
X. Wang, D.D. Kang, K. Shen, C. Song, S. Lu, L.C. Chang, S.G. Liao, Z. Huo, S. Tang, Y. Ding, N. Kaminski, E. Sibille, Y. Lin, J. Li, G.C. Tseng, An R package suite for microarray meta-analysis in quality control, differentially expressed gene analysis and pathway enrichment detection. Bioinformatics 28, 2534–2536 (2012). https://doi.org/10.1093/bioinformatics/bts485
G. Schwarzer, metacor: General package for meta-analysis (2016)
E. Laliberté, metacor: Meta-analysis of correlation coefficients (2011)
P. Langfelder, S. Horvath, WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). https://doi.org/10.1186/1471-2105-9-559
P. Shannon, A. Markiel, O. Ozier, N.S. Baliga, J.T. Wang, D. Ramage, N. Amin, B. Schwikowski, T. Ideker, Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). https://doi.org/10.1101/gr.1239303
A. Franceschini, D. Szklarczyk, S. Frankild, M. Kuhn, M. Simonovic, A. Roth, J. Lin, P. Minguez, P. Bork, C. von Mering, L.J. Jensen, STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013). https://doi.org/10.1093/nar/gks1094
R. Janky, A. Verfaillie, H. Imrichova, B. Van de Sande, L. Standaert, V. Christiaens, G. Hulselmans, K. Herten, M. Naval Sanchez, D. Potier, D. Svetlichnyy, Z. Kalender Atak, M. Fiers, J.C. Marine, S. Aerts, iRegulon: From a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 10, e1003731 (2014). https://doi.org/10.1371/journal.pcbi.1003731
W. da Huang, B.T. Sherman, R.A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). https://doi.org/10.1038/nprot.2008.211
A.D. Rouillard, G.W. Gundersen, N.F. Fernandez, Z. Wang, C.D. Monteiro, M.G. McDermott, A. Ma'ayan, The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016 (2016). https://doi.org/10.1093/database/baw100
E. Cerami, J. Gao, U. Dogrusoz, B.E. Gross, S.O. Sumer, B.A. Aksoy, A. Jacobsen, C.J. Byrne, M.L. Heuer, E. Larsson, Y. Antipin, B. Reva, A.P. Goldberg, C. Sander, N. Schultz, The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). https://doi.org/10.1158/2159-8290.CD-12-0095
M. Jia, T. Andreassen, L. Jensen, T.F. Bathen, I. Sinha, H. Gao, C. Zhao, L.A. Haldosen, Y. Cao, L. Girnita, S.A. Moestue, K. Dahlman-Wright, Estrogen receptor alpha promotes breast cancer by reprogramming choline metabolism. Cancer Res 76, 5634–5646 (2016). https://doi.org/10.1158/0008-5472.CAN-15-2910
Q. Li, N.J. Birkbak, B. Gyorffy, Z. Szallasi, A.C. Eklund, Jetset: Selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics 12, 474 (2011). https://doi.org/10.1186/1471-2105-12-474
K.R. Coser, J. Chesnes, J. Hur, S. Ray, K.J. Isselbacher, T. Shioda, Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci U S A 100, 13994–13999 (2003). https://doi.org/10.1073/pnas.2235866100
E. Guivarc'h, M. Buscato, A.L. Guihot, J. Favre, E. Vessieres, L. Grimaud, J. Wakim, N.J. Melhem, R. Zahreddine, M. Adlanmerini, Predominant role of nuclear versus membrane estrogen receptor α in arterial protection: Implications for estrogen receptor α modulation in cardiovascular prevention/safety. J Am Heart Assoc 7, e008950 (2018)
J. Fu, L. Bian, L. Zhao, Z. Dong, X. Gao, H. Luan, Y. Sun, H. Song, Identification of genes for normalization of quantitative real-time PCR data in ovarian tissues. Acta Biochim Biophys Sin 42, 568–574 (2010)
K. Stokes, B. Alston-Mills, C. Teng, Estrogen response element and the promoter context of the human and mouse lactoferrin genes influence estrogen receptor α-mediated transactivation activity in mammary gland cells. J Mol Endocrinol 33, 315–334 (2004)
B.H. Akman, T. Can, A.E. Erson-Bensan, Estrogen-induced upregulation and 3'-UTR shortening of CDC6. Nucleic Acids Res 40, 10679–10688 (2012). https://doi.org/10.1093/nar/gks855
H. Alotaibi, E.C. Yaman, E. Demirpençe, U.H. Tazebay, Unliganded estrogen receptor-α activates transcription of the mammary gland Na+/I− symporter gene. Biochem Biophys Res Commun 345, 1487–1496 (2006)
K.J. Livak, T.D. Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408 (2001)
A. Bahreini, Z. Li, P. Wang, K.M. Levine, N. Tasdemir, L. Cao, H.M. Weir, S.L. Puhalla, N.E. Davidson, A.M. Stern, D. Chu, B.H. Park, A.V. Lee, S. Oesterreich, Mutation site and context dependent effects of ESR1 mutation in genome-edited breast cancer cell models. Breast Cancer Res 19, 60–60 (2017). https://doi.org/10.1186/s13058-017-0851-4
D.C. Koboldt, R.S. Fulton, M.D. McLellan, H. Schmidt, J. Kalicki-Veizer, J.F. McMichael, L.L. Fulton, D.J. Dooling, L. Ding, E.R. Mardis, R.K. Wilson, A. Ally, M. Balasundaram, Y.S.N. Butterfield, R. Carlsen, C. Carter, A. Chu, E. Chuah, H.-J.E. Chun, R.J.N. Coope, N. Dhalla, R. Guin, C. Hirst, M. Hirst, R.A. Holt, D. Lee, H.I. Li, M. Mayo, R.A. Moore, A.J. Mungall, E. Pleasance, A.G. Robertson, J.E. Schein, A. Shafiei, P. Sipahimalani, J.R. Slobodan, D. Stoll, A. Tam, N. Thiessen, R.J. Varhol, N. Wye, T. Zeng, Y. Zhao, I. Birol, S.J.M. Jones, M.A. Marra, A.D. Cherniack, G. Saksena, R.C. Onofrio, N.H. Pho, S.L. Carter, S.E. Schumacher, B. Tabak, B. Hernandez, J. Gentry, H. Nguyen, A. Crenshaw, K. Ardlie, R. Beroukhim, W. Winckler, G. Getz, S.B. Gabriel, M. Meyerson, L. Chin, P.J. Park, R. Kucherlapati, K.A. Hoadley, J.T. Auman, C. Fan, Y.J. Turman, Y. Shi, L. Li, M.D. Topal, X. He, H.-H. Chao, A. Prat, G.O. Silva, M.D. Iglesia, W. Zhao, J. Usary, J.S. Berg, M. Adams, J. Booker, J. Wu, A. Gulabani, T. Bodenheimer, A.P. Hoyle, J.V. Simons, M.G. Soloway, L.E. Mose, S.R. Jefferys, S. Balu, J.S. Parker, D.N. Hayes, C.M. Perou, S. Malik, S. Mahurkar, H. Shen, D.J. Weisenberger, T. Triche Jr., P.H. Lai, M.S. Bootwalla, D.T. Maglinte, B.P. Berman, D.J. Van Den Berg, S.B. Baylin, P.W. Laird, C.J. Creighton, L.A. Donehower, G. Getz, M. Noble, D. Voet, G. Saksena, N. Gehlenborg, D. DiCara, J. Zhang, H. Zhang, C.-J. Wu, S.Y. Liu, M.S. Lawrence, L. Zou, A. Sivachenko, P. Lin, P. Stojanov, R. Jing, J. Cho, R. Sinha, R.W. Park, M.-D. Nazaire, J. Robinson, H. Thorvaldsdottir, J. Mesirov, P.J. Park, L. Chin, S. Reynolds, R.B. Kreisberg, B. Bernard, R. Bressler, T. Erkkila, J. Lin, V. Thorsson, W. Zhang, I. Shmulevich, G. Ciriello, N. Weinhold, N. Schultz, J. Gao, E. Cerami, B. Gross, A. Jacobsen, R. Sinha, B.A. Aksoy, Y. Antipin, B. Reva, R. Shen, B.S. Taylor, M. Ladanyi, C. Sander, P. Anur, P.T. Spellman, Y. Lu, W. Liu, R.R.G. Verhaak, G.B. Mills, R. Akbani, N. Zhang, B.M. Broom, T.D. Casasent, C. Wakefield, A.K. Unruh, K. Baggerly, K. Coombes, J.N. Weinstein, D. Haussler, C.C. Benz, J.M. Stuart, S.C. Benz, J. Zhu, C.C. Szeto, G.K. Scott, C. Yau, E.O. Paull, D. Carlin, C. Wong, A. Sokolov, J. Thusberg, S. Mooney, S. Ng, T.C. Goldstein, K. Ellrott, M. Grifford, C. Wilks, S. Ma, B. Craft, C. Yan, Y. Hu, D. Meerzaman, J.M. Gastier-Foster, J. Bowen, N.C. Ramirez, A.D. Black, R.E. Pyatt, P. White, E.J. Zmuda, J. Frick, T.M. Lichtenberg, R. Brookens, M.M. George, M.A. Gerken, H.A. Harper, K.M. Leraas, L.J. Wise, T.R. Tabler, C. McAllister, T. Barr, M. Hart-Kothari, K. Tarvin, C. Saller, G. Sandusky, C. Mitchell, M.V. Iacocca, J. Brown, B. Rabeno, C. Czerwinski, N. Petrelli, O. Dolzhansky, M. Abramov, O. Voronina, O. Potapova, J.R. Marks, W.M. Suchorska, D. Murawa, W. Kycler, M. Ibbs, K. Korski, A. Spychała, P. Murawa, J.J. Brzeziński, H. Perz, R. Łaźniak, M. Teresiak, H. Tatka, E. Leporowska, M. Bogusz-Czerniewicz, J. Malicki, A. Mackiewicz, M. Wiznerowicz, X. Van Le, B. Kohl, N.V. Tien, R. Thorp, N. Van Bang, H. Sussman, B.D. Phu, R. Hajek, N.P. Hung, T.V.T. Phuong, H.Q. Thang, K.Z. Khan, R. Penny, D. Mallery, E. Curley, C. Shelton, P. Yena, J.N. Ingle, F.J. Couch, W.L. Lingle, T.A. King, A.M. Gonzalez-Angulo, G.B. Mills, M.D. Dyer, S. Liu, X. Meng, M. Patangan, N. The Cancer Genome Atlas, L. Genome sequencing centres: Washington University in St, B.C.C.A. Genome characterization centres, I. Broad, Brigham, H. Women’s, S. Harvard Medical, C.H. University of North Carolina, H. University of Southern California/Johns, M. Genome data analysis: Baylor College of, B. Institute for Systems, C. Memorial Sloan-Kettering Cancer, H. Oregon, U. Science, M.D.A.C.C. The University of Texas, S.C.B.I. University of California, Nci, R. Biospecimen core resource: Nationwide Children’s Hospital Biospecimen Core, A.-I. Tissue source sites, Christiana, Cureline, C. Duke University Medical, C. The Greater Poland Cancer, Ilsbio, C. International Genomics, C. Mayo, Mskcc and M.D.A.C. Center, Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). https://doi.org/10.1038/nature11412
T.J. Robinson, J.C. Liu, F. Vizeacoumar, T. Sun, N. Maclean, S.E. Egan, A.D. Schimmer, A. Datti, E. Zacksenhaus, RB1 status in triple negative breast cancer cells dictates response to radiation treatment and selective therapeutic drugs. PLoS One 8, e78641 (2013). https://doi.org/10.1371/journal.pone.0078641
P. Russo, A. Del Bufalo, M. Milic, G. Salinaro, M. Fini, A. Cesario, Cholinergic receptors as target for cancer therapy in a systems medicine perspective. Curr Mol Med 14, 1126–1138 (2014)
J.J. Pink, V.C. Jordan, Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res 56, 2321–2330 (1996)
M.C. Alles, M. Gardiner-Garden, D.J. Nott, Y. Wang, J.A. Foekens, R.L. Sutherland, E.A. Musgrove, C.J. Ormandy, Meta-analysis and gene set enrichment relative to er status reveal elevated activity of MYC and E2F in the "basal" breast cancer subgroup. PLoS One 4, e4710 (2009). https://doi.org/10.1371/journal.pone.0004710
L. Verlinden, I.V. Bempt, G. Eelen, M. Drijkoningen, I. Verlinden, K. Marchal, C. De Wolf-Peeters, M.R. Christiaens, L. Michiels, R. Bouillon, A. Verstuyf, The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor−/progesterone receptor-/HER-2- breast carcinomas. Cancer Res 67, 6574–6581 (2007). https://doi.org/10.1158/0008-5472.CAN-06-3545
C.-H. Lin, H.-H. Lee, C.-H. Kuei, H.-Y. Lin, L.-S. Lu, F.-P. Lee, J. Chang, J.-Y. Wang, K.-C. Hsu, Y.-F. Lin, Nicotinic acetylcholine receptor subunit Alpha-5 promotes radioresistance via recruiting E2F activity in oral squamous cell carcinoma. J Clin Med 8, 1454 (2019)
W. Wang, L. Dong, B. Saville, S. Safe, Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Sp1/estrogen receptor interactions. Mol Endocrinol 13, 1373–1387 (1999). https://doi.org/10.1210/mend.13.8.0323
K. Dahlman-Wright, Y. Qiao, P. Jonsson, J. Gustafsson, C. Williams, C. Zhao, Interplay between AP-1 and estrogen receptor α in regulating gene expression and proliferation networks in breast cancer cells. Carcinogenesis 33, 1684–1691 (2012). https://doi.org/10.1093/carcin/bgs223
J. Lukas, B.O. Petersen, K. Holm, J. Bartek, K. Helin, Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol Cell Biol 16, 1047–1057 (1996). https://doi.org/10.1128/mcb.16.3.1047
H. Singhal, M.E. Greene, A.L. Zarnke, M. Laine, R. Al Abosy, Y.F. Chang, A.G. Dembo, K. Schoenfelt, R. Vadhi, X. Qiu, P. Rao, B. Santhamma, H.B. Nair, K.J. Nickisch, H.W. Long, L. Becker, M. Brown, G.L. Greene, Progesterone receptor isoforms, agonists and antagonists differentially reprogram estrogen signaling. Oncotarget 9, 4282–4300 (2018). https://doi.org/10.18632/oncotarget.21378
R. Mahadevappa, H. Neves, S.M. Yuen, Y. Bai, C.M. McCrudden, H.F. Yuen, Q. Wen, S.D. Zhang, H.F. Kwok, The prognostic significance of Cdc6 and Cdt1 in breast cancer. Sci Rep 7, 985 (2017). https://doi.org/10.1038/s41598-017-00998-9
G. Nassa, R. Tarallo, G. Giurato, M.R. De Filippo, M. Ravo, F. Rizzo, C. Stellato, C. Ambrosino, M. Baumann, N. Lietzèn, T.A. Nyman, A. Weisz, Post-transcriptional regulation of human breast cancer cell proteome by unliganded estrogen receptor β via microRNAs. Mol Cell Proteomics 13, 1076–1090 (2014). https://doi.org/10.1074/mcp.M113.030403
E.V. Jensen, G. Cheng, C. Palmieri, S. Saji, S. Mäkelä, S. Van Noorden, T. Wahlström, M. Warner, R.C. Coombes, J.A. Gustafsson, Estrogen receptors and proliferation markers in primary and recurrent breast cancer. Proc Natl Acad Sci U S A 98, 15197–15202 (2001). https://doi.org/10.1073/pnas.211556298
U.K. Mukhopadhyay, C.C. Oturkar, C. Adams, N. Wickramasekera, S. Bansal, R. Medisetty, A. Miller, W.M. Swetzig, L. Silwal-Pandit, A.-L. Børresen-Dale, C.J. Creighton, J.H. Park, S.D. Konduri, A. Mukhopadhyay, A. Caradori, A. Omilian, W. Bshara, B.A. Kaipparettu, G.M. Das, TP53 status as a determinant of pro- vs anti-tumorigenic effects of estrogen receptor-beta in breast cancer. J Natl Cancer Inst 111, 1202-1215 (2019). https://doi.org/10.1093/jnci/djz051
S.K. Gruvberger-Saal, P.-O. Bendahl, L.H. Saal, M. Laakso, C. Hegardt, P. Edén, C. Peterson, P. Malmström, J. Isola, Å. Borg, Estrogen receptor β expression is associated with tamoxifen response in ERα-negative breast carcinoma. Clin Cancer Res 13, 1987–1994 (2007)
P. O'neill, M. Davies, A. Shaaban, H. Innes, A. Torevell, D. Sibson, C. Foster, Wild-type oestrogen receptor beta (ERβ1) mRNA and protein expression in Tamoxifen-treated post-menopausal breast cancers. Br J Cancer 91, 1694–1702 (2004)
R. Girgert, G. Emons, C. Gründker, Estrogen signaling in ERα-negative breast cancer: ERβ and GPER. Front Endocrinol 9, 781 (2018)
L.-H. Hsu, N.-M. Chu, Y.-F. Lin, S.-H. Kao, G-protein coupled estrogen receptor in breast cancer. Int J Mol Sci 20, 306 (2019)
L. Salvatori, L. Ravenna, F. Caporuscio, L. Principessa, G. Coroniti, L. Frati, M.A. Russo, E. Petrangeli, Action of retinoic acid receptor on EGFR gene transactivation and breast cancer cell proliferation: Interplay with the estrogen receptor. Biomed Pharmacother 65, 307–312 (2011)
V.C. Jordan, R. Curpan, P.Y. Maximov, Estrogen receptor mutations found in breast cancer metastases integrated with the molecular pharmacology of selective ER modulators. J Natl Cancer Inst 107, djv075 (2015)
F.J. Fleming, A.D. Hill, E.W. McDermott, N.J. O’Higgins, L.S. Young, Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by β-estradiol and 4-hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metabol 89, 375–383 (2004)
E.N. Lyukmanova, M.L. Bychkov, G.V. Sharonov, A.V. Efremenko, M.A. Shulepko, D.S. Kulbatskii, Z.O. Shenkarev, A.V. Feofanov, D.A. Dolgikh, M.P. Kirpichnikov, Human secreted proteins SLURP-1 and SLURP-2 control the growth of epithelial cancer cells via interactions with nicotinic acetylcholine receptors. Br J Pharmacol 175, 1973–1986 (2018). https://doi.org/10.1111/bph.14194
A.R. Salem, P. Martínez Pulido, F. Sanchez, Y. Sanchez, A.J. Español, M.E. Sales, Effect of low dose metronomic therapy on MCF-7 tumor cells growth and angiogenesis. Role of muscarinic acetylcholine receptors Int Immunopharmacol 84, 106514 (2020). https://doi.org/10.1016/j.intimp.2020.106514
T.I. Terpinskaya, A.V. Osipov, T.V. Balashevich, T.L. Yanchanka, E.A. Tamashionik, V.I. Tsetlin, Y.N. Utkin, Blockers of nicotinic acetylcholine receptors delay tumor growth and increase antitumor activity of mouse splenocytes. Dokl Biochem Biophys 491, 89–92 (2020). https://doi.org/10.1134/S1607672920020143
Z. Sun, J. Bao, M. Zhangsun, S. Dong, D. Zhangsun, S. Luo, αO-Conotoxin GeXIVA inhibits the growth of breast cancer cells via interaction with α9 nicotine acetylcholine receptors. Marine drugs 18, 195 (2020). https://doi.org/10.3390/md18040195
S.A. Ochsner, D.L. Steffen, S.G. Hilsenbeck, E.S. Chen, C. Watkins, N.J. McKenna, GEMS (gene expression MetaSignatures), a web resource for querying meta-analysis of expression microarray datasets: 17beta-estradiol in MCF-7 cells. Cancer Res 69, 23–26 (2009). https://doi.org/10.1158/0008-5472.CAN-08-3492
D.R. Rhodes, J. Yu, K. Shanker, N. Deshpande, R. Varambally, D. Ghosh, T. Barrette, A. Pandey, A.M. Chinnaiyan, Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A 101, 9309–9314 (2004). https://doi.org/10.1073/pnas.0401994101
B.J. Wilson, V. Giguere, Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway. Mol Cancer 7, 49 (2008). 10.1186/1476–4598-7-49
D.D. Smith, P. Saetrom, O. Snove Jr., C. Lundberg, G.E. Rivas, C. Glackin, G.P. Larson, Meta-analysis of breast cancer microarray studies in conjunction with conserved cis-elements suggest patterns for coordinate regulation. BMC Bioinformatics 9, 63 (2008). https://doi.org/10.1186/1471-2105-9-63
Acknowledgments
This study was funded by a research grant from The Scientific and Technological Research Council of Turkey (to OK; TUBITAK, 111 T316). We are thankful to the Higher Education Commission (HEC), Pakistan for funding Huma Shehwana for her PhD studies. We thank TUBITAK for financial assistance to Emine Sila Ozdemir during her Master studies (Scholarship 2210-E). We also thank Can Alkan and Marzieh Eslami Rasekh for helping us with the computation power needed to process microarray files.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shehwana, H., Keskus, A.G., Ozdemir, S.E. et al. CHRNA5 belongs to the secondary estrogen signaling network exhibiting prognostic significance in breast cancer. Cell Oncol. 44, 453–472 (2021). https://doi.org/10.1007/s13402-020-00581-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-020-00581-x