Abstract
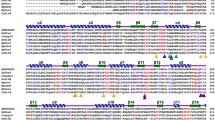
Despite extensive use in the biofuel industry, only butyryl co-A dehydrogenase enzymes from the Clostridia group have undergone extensive structural and genetic characterization. The present study, portrays the characterization of structural, functional and phylogenetic properties of butyryl co-A dehydrogenase identified within the genome of Pusillimonas ginsengisoli SBSA. In silico characterization, homology modelling and docking data indicates that this protein is a homo-tetramer and 388 amino acid residue long, rich in alanine and leucine residue; having molecular weight of 42347.69 dalton. Its isoelectric point value is 5.78; indicate its neutral nature while 38.38 instability index value indicate its stable nature. Its thermostable nature evidenced by its high aliphatic index (93.14); makes its suitable for industry-based use. The secondary structure prediction analysis of butyryl co-A dehydrogenase unveiled that the proteins has secondary arrangements of 54% α-helix, 13% β-stand and 5% disordered conformation. However, phylogenetic analysis clearly indicates that probably horizontal gene transfer is the primary mechanism of spreading of this gene in this organism. Notably, multiple sequence alignment study of phylogenetically diverse butyryl co-A dehydrogenase sequence highlighted the presence of conserved amino acid residues i.e. YXV/LGXKXWXS/T. Physicochemical characterization of other relevant proteins involved in butanol metabolism of SBSA also has been carried out. However, metabolic construction of functional butanol biosynthesis pathway in SBSA, enlightened its cost-effective potential use in biofuel industry as an alternate to Clostridia system.





Similar content being viewed by others
References
Andorf C, Dobbs D, Honavar V (2007) Exploring inconsistencies in genome-wide protein function annotations: a machine learning approach. BMC Bioinform 8:284. https://doi.org/10.1186/1471-2105-8-284
Atsumi S, Liao J (2008) Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotechnol 19:414–419. https://doi.org/10.1016/j.copbio.2008.08.008
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. https://doi.org/10.1093/bioinformatics/btq662
Bennett GN, Rudolph FB (1995) The central metabolic pathway from acetyl-CoA to butyryl-CoA in Clostridium acetobutylicum. FEMS Microbiol Rev 17:241–249. https://doi.org/10.1111/j.1574-6976.1995.tb00208.x
Berezina OV, Zakharova NV, Brandt A et al (2010) Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl Microbiol Biotechnol 87:635–646. https://doi.org/10.1007/s00253-010-2480-z
Berezina OV, Zakharova NV, Yarotsky CV et al (2012) Microbial producers of butanol. Appl Biochem 48:625–638. https://doi.org/10.1134/S0003683812070022
Berman HM, Westbrook J, Feng Z et al (2000) RCSB Protein Data Bank: Structural biology views for basic and applied research. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42(W1):W252–W258
Bramucci MG, Eliot AC, Maggio-Hall LA, Nakamura CE, Butamax Advanced Biofuels LLC (2012) Butanol dehydrogenase enzyme from the bacterium Achromobacter xylosoxidans. U.S. Patent 8,188,250
Bramucci MG, Eliot AC, Maggio-Hall LA, Nakamura CE (2014) Butamax Advanced Biofuels LLC, Butanol dehydrogenase enzyme from the bacterium Achromobacter xylosoxidans. U.S. Patent 8,691,540.
Caspi R, Foerster H, Fulcher CA et al (2006) MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res 34:D511–D516. https://doi.org/10.1093/nar/gkj128
Chou PY, Fasman GD (1974) Conformational parameters for amino acids in helical, β-sheet, and random coil regions calculated from proteins. Biochemistry 13:211–222. https://doi.org/10.1021/bi00699a001
Chou PY, Fasman GD (1974) Prediction of protein conformation. Biochemistry 13:222–245. https://doi.org/10.1021/bi00699a002
Discovery studio (2017). R2 BIOVIA, San Diego: Dassault Systèmes, 2017. http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization.html
Djordjevic S, Pace CP, Stankovich MT et al (1995) Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera elsdenii. Biochemistry 34:2163–2171
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Fox AR, Soto G, Mozzicafreddo M et al (2014) Understanding the function of bacterial and eukaryotic thiolases II by integrating evolutionary and functional approaches. Gene 533:5–10. https://doi.org/10.1016/j.gene.2013.09.096
Francke C, Siezen RJ, Teusink B (2005) Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol 13:550–558. https://doi.org/10.1016/j.tim.2005.09.001
Gasteiger E, Hoogland C, Gattiker A et al (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The Proteomics Protocols Handbook, Springer Protocols Handbooks. Humana Press, Totowa. https://doi.org/10.1385/1-59259-890-0:571
Geourjon C, Deléage G (1995) SOPMA: significant improvements in proteinsecondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11:681–684. https://doi.org/10.1093/bioinformatics/11.6.681
Green EM (2011) Fermentative production of butanol—the industrial perspective. Curr Opin Biotechnol 22:337–343. https://doi.org/10.1016/j.copbio.2011.02.004
Guruprasad K, Reddy BB, Pandit MW (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng Des Sel 4:155–161. https://doi.org/10.1093/protein/4.2.155
Huang H, Liu H, Gan YR (2010) Genetic modification of critical enzymes and involved genes in butanol biosynthesis from biomass. Biotechnol Adv 28:651–657. https://doi.org/10.1016/j.biotechadv.2010.05.015
Hu B, Lidstrom ME (2014) Metabolic engineering of Methylobacterium extorquens AM1 for 1-butanol production. Biotechnol Biofuels 7:156. https://doi.org/10.1186/s13068-014-0156-0
Ikai A (1980) Thermostability and aliphatic index of globular proteins. J Biochem 88:1895–1898. https://doi.org/10.1093/oxfordjournals.jbchem.a133168
Islam K, Pal K, Debnath U, Sidick Basha R, Khan AT, Jana K, Misra AK (2020) Anti-cancer potential of (1,2-dihydronaphtho[2,1-b]furan-2-yl)methanone derivatives. Bioorg Med Chem Lett. https://doi.org/10.1016/j.bmcl.2020.127476
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From Appl Microbiol Biotechnol (2008) 80:849–862 861 genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354–D357
Kataoka N, Tajima T, Kato J et al (2011) Development of butanol-tolerant Bacillus subtilis strain GRSW2-B1 as a potential bioproduction host. AMB Express 1:10. https://doi.org/10.1186/2191-0855-1-10
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Kim EJ, Kim YJ, Kim KJ et al (2014) Structural insights into substrate specificity of crotonase from the n-butanol producing bacterium Clostridium acetobutylicum. Biochem Biophys Res Commun 451:431–435. https://doi.org/10.1016/j.bbrc.2014.07.139
Kishino H, Hasegawa M (2001) Maximum likelihood. Encycl Genet. https://doi.org/10.1006/RWGN.2001.0803
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kurtz S, Phillippy A, Delcher AL et al (2004) Versatile and open software for comparing large genomes. Genome Biol 5:12. https://doi.org/10.1186/gb-2004-5-2-r12
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. https://doi.org/10.1016/0022-2836(82)90515-0
Lan EI, Liao JC (2011) Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab Eng 13:353–363. https://doi.org/10.1016/j.ymben.2011.04.004
Loder AJ, Zeldes BM, Garrison GD et al (2015) Alcohol selectivity in a synthetic thermophilic n-butanol pathway is driven by biocatalytic and thermostability characteristics of constituent enzymes. Appl Environ Microbiol 81:7187–7200. https://doi.org/10.1128/AEM.02028-15
Lovell SC, Davis IW, Arendall WB et al (2003) Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437e450. https://doi.org/10.1002/prot.10286
Lütke-Eversloh T, Bahl H (2011) Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr Opin Biotechnol 22:634–647. https://doi.org/10.1016/j.copbio.2011.01.011
Mandal S, Rameez MJ, Chatterjee S et al (2020) Molecular mechanism of sulfur chemolithotrophy in the betaproteobacterium Pusillimonas ginsengisoli SBSA. Microbiology 166:386–397. https://doi.org/10.1099/mic.0.000890
Marchler-Bauer A, Lu S, Anderson JB et al (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. https://doi.org/10.1093/nar/gkq1189
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. https://doi.org/10.1093/bioinformatics/16.4.404
Moldoveanu SC, David V, Moldoveanu SC et al (2017) Properties of analytes and matrices determining HPLC Selection. Sel HPLC Method Chem Anal. https://doi.org/10.1016/B978-0-12-803684-6.00005-6
Moon HG, Jang YS, Cho C, Lee J, Binkley R, Lee SY (2016) One hundred years of clostridial butanol fermentation. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fnw001
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Müller N, Schleheck D, Schink B (2009) Involvement of NADH: acceptor oxidoreductase and butyryl coenzyme A dehydrogenase in reversed electron transport during syntrophic butyrate oxidation by Syntrophomonas wolfei. J. Bacteriol 191:6167–6177. https://doi.org/10.1128/JB.01605-08
Nei M, Zhang J (2006) Evolutionary distance: estimation. In: Encyclopedia of life sciences. Wiley, Chichester.
Reynolds CR, Islam SA, Sternberg MJE (2018) EzMol: A web server wizard for the rapid visualisation and image production of protein and nucleic acid structures. J Mol Biol 430:2244–8
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277. https://doi.org/10.1016/S0168-9525(00)02024-2
Schmidt A, Müller N, Schink B et al (2013) A proteomic view at the biochemistry of syntrophic butyrate oxidation in Syntrophomonas wolfei. PLoS ONE. https://doi.org/10.1371/journal.pone.0056905
Schnoes AM, Brown SD, Dodevski I, Babbitt PC (2009) Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol 5:e1000605. https://doi.org/10.1371/journal.pcbi.1000605
Seeliger D, De Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24:417–422. https://doi.org/10.1007/s10822-010-9352-6
Sillers R, Al-Hinai MA, Papoutsakis ET (2009) Aldehyde–alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase down regulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicum fermentations. Biotechnol. Bioeng 102:38–49. https://doi.org/10.1002/bit.22058
Stim-Herndon KP, Petersen DJ, Bennett GN (1995) Characterization of an acetyl-CoA C-acetyltransferase (thiolase) gene from Clostridium acetobutylicum ATCC 824. Gene 154:81–85. https://doi.org/10.1016/0378-1119(94)00838-J
Tanaka Y, Kasahara K, Hirose Y et al (2017) Enhancement of butanol production by sequential introduction of mutations conferring butanol tolerance and streptomycin resistance. J Biosci Bioeng 124:400–407. https://doi.org/10.1016/j.jbiosc.2017.05.003
Williamson G, Engel PC (1984) Butyryl-CoA dehydrogenase from Megasphaera elsdenii. Specificity of the catalytic reaction. Biochem J 218:521–529
Wong BJ, Gerlt JA (2004) Evolution of function in the crotonase superfamily:(3 S)-methylglutaconyl-CoA hydratase from Pseudomonas putida. Biochemistry 43:4646–4654. https://doi.org/10.1021/bi0360307
Xu Y, Li H, Jin YH et al (2014) Dimerization interface of 3-hydroxyacyl-CoA dehydrogenase tunes the formation of its catalytic intermediate. PloS one, 9(4). doi: https://doi.org/10.1371/journal.pone.0095965.
Yadav PK, Singh G, Gautam B et al (2013) Molecular modeling, dynamics studies and virtual screening of Fructose 1, 6 biphosphate aldolase-II in community acquired-methicillin resistant Staphylococcus aureus (CA-MRSA). Bioinformation 9:158. https://doi.org/10.6026/97320630009158
Zhang J, Yu L, Lin M et al (2017) n-Butanol production from sucrose and sugarcane juice by engineered Clostridium tyrobutyricum overexpressing sucrose catabolism genes and adhE2. Bioresour Technol 233:51–57. https://doi.org/10.1016/j.biortech.2017.02.079
Zheng YN, Li LZ, Xian M, Ma YJ, Yang JM, Xu X, He DZ (2009) Problems with the microbial production of butanol. J Ind Microbiol Biot 36:1127–1138
Acknowledgements
We thank Dr Wriddhiman Ghosh (Bose Institute, India) for his helpful suggestions in manuscript preparation and for providing all kind of computational facility. JS received fellowships from Council of Scientific and Industrial Research, GoI.
Author information
Authors and Affiliations
Contributions
SM conceived the study, designed the experiments, interpreted the results and wrote the paper. SM, UD and JS performed bioinformatics analysis. All authors read and vetted the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Kerry Geiler-Samerotte.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mandal, S., Debnath, U. & Sarkar, J. Structural-Genetic Characterization Of Novel Butaryl co-A Dehydrogenase and Proposition of Butanol Biosynthesis Pathway in Pusillimonas ginsengisoli SBSA. J Mol Evol 89, 81–94 (2021). https://doi.org/10.1007/s00239-020-09989-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-020-09989-3