Abstract
Phosphorus (P) deficiency is one of the major nutrient stresses restricting plant growth. The uptake of P by plants from soil is mainly mediated by the phosphate (Pi) transporters belonging to the PHT1 family. Multiple PHT1 genes from diverse plant species have been shown to be strongly up-regulated upon Pi starvation, however, the underlying mechanisms for the Pi-starvation-induced (PSI) up-regulation have not been well deciphered for most Pi transporter genes. Here, we reported a detailed dissection of the promoter activity of a PSI rice Pi transporter gene OsPT6, using the β-glucuronidase (GUS) reporter gene. OsPT6 promoter could drive GUS expression strongly in both roots and blades of rice plants grown under low P, but not high P. Cis-acting element analysis identified one copy of the P1BS motif and two copies of the W-box motif in OsPT6 promoter. Targeted deletion of the P1BS motif caused almost complete abolition of GUS induction in response to Pi starvation, irrespective of the presence or absence of the W-box motif, Four repeats of the P1BS motif fused to the CaMV35S minimal promoter was sufficient to induce GUS expression responsive to Pi starvation. Targeted deletion of the upstream W-box motif (W1) did not affect the GUS expression activity compared with the full-length OsPT6 promoter, while targeted deletion of the downstream W-box motif (W2) or both of the W-box motifs remarkably reduced the GUS induction rate upon Pi starvation. Our results proposed that the PSI response of OsPT6 was positively regulated by at least two elements, the sole P1BS and the downstream W-box, in its promoter, and the W-box-mediated up-regulation of OsPT6 might be highly dependent on the P1BS motif.
Similar content being viewed by others
Introduction
Phosphorus (P) is one of the most essential macro-nutrients for plant growth and development. It plays critical roles in numerous physiological and metabolic processes, including biosynthesis of macromolecules, energy transfer, signal transduction, photosynthesis and respiration (Clarkson et al. 2003). The primary source of P taken up by plants from soil is inorganic orthophosphate (Pi). As a result of chemical and biological fixations and the slow diffusion rate in soil, P is one of the least available plant nutrients in many nutrient-poor soils (Vance et al. 2003). To cope with P limiting stress, plants have evolved a series of subtle strategies to enhance P acquisition, including alterations in root morphology, formation of symbiotic associations with beneficial microbes and activation of a series of Pi-starvation-induced (PSI) genes (Lambers et al. 2015).
The Pi concentration in the rhizosphere rarely exceeds 10 μM, while plants need to maintain their cytoplasmic Pi concentrations at a millimolar range (Ragnothama et al. 1999). This indicates the requirement of specialized transport systems for the uptake of Pi and for its redistribution within plants. Transport of Pi through plant membranes is mediated by a number of transporters belonging to several families. Most of the Pi transporters identified thus far are typical of H + /Pi symporters, and could be grouped into the so-called high-affinity Pht1 family (Wang et al. 2018; Lin et al. 2009; Hermando et al. 2018). In the typical model plant species, Arabidopsis and rice, a total of 9 and 13 distinct Pht1 genes have been annotated, respectively, and most of them have been shown to be inducible at least at the transcription level by Pi starvation (Muchhal et al. 1996; Ai et al. 2009; Jia et al. 2011; Sun et al. 2012; Wu et al. 2013; Wang et al. 2014a, b; Lapis-Gaza et al. 2014; Karthikeyan et al. 2009; Nagarajan et al. 2011; Liu et al. 2010; Li et al. 2015).
Tissue/cellular distribution analysis indicated that members of the Pht1 family in many species are differentially expressed and divergent in function during plant development (Nussaume et al. 2011). The relatively high expression levels of some Pht1 members in roots, particularly in lateral root and root hairs well supports a role of these genes, such as AtPT1 and AtPT4, in Pi capture and uptake, especially under Pi deficiency conditions (Ye et al. 2015; Shin et al. 2004; Misson et al. 2004). In rice, OsPT6 was shown to be expressed in both epidermal and cortical cells of the young primary and lateral roots, and its protein was able to complement a yeast Pi uptake mutant in the high concentration range. knock-down of OsPT6 expression caused significant decrease in both the uptake and the long-distance transport of Pi from roots to shoots (Ai et al. 2009). The expression of some Pht1 genes, such as the rice OsPT1 and OsPT8, are ubiquitously distributed throughout plant tissues and showed no conspicuous response at transcriptional level to the alterations of external Pi status, suggesting a role of these Pht1 members in mediating the internal Pi redistribution (Jia et al. 2011; Li et al. 2015). Besides the Pi-responsive Pht1 genes, a growing number of Pi transporters genes of the Pht1 family have been identified to be specifically induced in roots colonized by arbuscular mycorrhizal (AM) fungi, and their functions have been proposed to be involved in Pi transport at the intraradical symbiotic interface (Sawers et al. 2017; Ferrol et al. 2019).
Although an increasing number of Pht1 genes associated with Pi signaling have been functionally characterized, the regulatory mechanisms underlying the PSI response have not been well deciphered for most of these Pht1 genes. Previous studies have identified multiple transcription factors, such as AtPHR1, OsPHR2, OsPTF1, AtWRKY75, OsWRKY74 and OsMYB1, probably involved in the regulation of Pi signaling or homeostasis in plants (Guo et al. 2015; Rubio et al. 2001; Yi et al. 2005; Devaiah et al. 2007; Zhou et al. 2008; Dai et al. 2016; Gu et al. 2017), however, to date, only a very few cis-acting elements, such as P1BS (PHR1-binding sequence) and W-box, have been experimentally validated to mediate PSI expression (Rubio et al. 2001; Bustos et al. 2010; Wang et al. 2014a, b). In our previous study, we reported a rice Pht1 gene, OsPT6, that was strongly up-regulated in both roots and blades upon Pi starvation (Ai et al. 2009; Li et al. 2014). The aim of this study was to identify the potential cis-acting elements in its promoter, thus facilitating the isolation and functional characterization of the transcription factor(s) responsible for the activation of the PSI response in future work. Fine analysis of the OsPT6 promoter by successive truncation and targeted deletion fused to a GUS reporter revealed that the up-regulation of OsPT6 in response to Pi starvation was positively regulated by at least two conserved elements, the P1BS and W-box, and the W-box-mediated regulation of PSI response of OsPT6 might be highly dependent on the P1BS motif.
Materials and methods
Plant materials and cultivation conditions
The rice (Oryza sativa ssp japonica) transgenic plants used in this study was in the cv. Nipponbare background. Seeds of the T2 independent transgenic lines were surface sterilized with 10% (v/v) sodium hypochlorite for 10 min and then germinated in a growth chamber programmed for 16 h of light at 30 °C and 8 h of dark at 22 °C. Ten-day-old seedlings were transferred to hydroponic culture containing the following nutrients: 1.25 mM NH4NO3, 0.35 mM K2SO4, 1 mM CaCl2, 1 mM MgSO4, 0.5 mM Na2SiO3, 20 µM Fe-EDTA, 20 µM H3BO3, 9 µM MnCl2, 0.32 µM CuSO4, 0.77 µM ZnSO4, and 0.39 µM Na2MoO4, supplemented with either 300 µM Pi as a high-P (HP) treatment or 15 µM Pi as a low-P (LP) treatment The nutrient solution was refreshed every 3 days.
For transgenic tobacco plants, seeds were surface sterilized in a 30% (v/v) H2O2 solution for 30 min, and then germinated and grown on MS medium containing either limited (10 µM) or sufficient (1 mM) Pi. Samples were collected after the plants were treated for 3 weeks.
Construction of the OsPT6 promoter binary vector
A series of promoter deletions of OsPT6 immediately upstream of the translation start ATG were amplified by PCR using specific primers listed in Table S2, and then introduced into the pS1aGUS-3 binary vector to replace the CaMV 35S promoter in front of the GUS reporter gene (Li et al. 2014; Ai et al. 2009). According to the length of different promoter fragments, the resulting constructs were designated as P-n respectively. The targeted deletion of P1BS or W-box was generated by overlap and extension PCR with the Fast Mutagenesis Kit V2 (Vazyme, Nanjing, China). To generate the 4XP1BS-GUS construct, four tandem repeats of the P1BS motif (GGATATTC) from the OsPT6 promoter was synthesized and fused to the 35S minimal promoter. The chimeric promoter fragment was subsequently cloned into the pCAMBIA1300 binary vector in front of the GUS reporter gene. These new derived binary vectors were introduced into Agrobacterium tumefaciens strain EHA105 for transformation of rice (Oryza sativa ssp japonica) or tobacco (Nicotiana tabacum L.) plants.
Plant transformation
Agrobacterium-mediated transformation of rice plants was performed as described previously (Upadhyaya et al. 2000).The transformation of tobacco plants was carried out using the leaf disk infection method (Fraley et al. 1983). The screening of positive transgenic plants were performed by using PCR assay. At least 10 independent transgenic lines were produced for each construct and tested by GUS histochemical assay.
Histochemical GUS assays
Histochemical localization of GUS expression was performed as described previously (Li et al. 2015). Excised samples were vacuum infiltrated with staining solution and incubated overnight at 37 ℃. Chlorophyll was removed by submerging the tissue in 70% ethanol. Representative plant materials were photographed using OLYMPUS MVX10 stereomicroscope. Fluorometric assays of GUS activities were determined according to (Li et al. 2014). Samples were homogenized in GUS extraction buffer (50 mM NaPO4, pH 7.0, 10 mM 2-mercaptoethanol, 10 mM Na2-EDTA, 0.1% sodium dodecylsulphate, 0.1% Triton X-100). GUS enzymatic activity was calculated as nmol 4-Methylumbelliferone (4-MU) per hour per microgram protein. For each construct, ten independent assays were performed and the average was calculated.
Quantitative real-time RT-PCR (qRT-PCR)
Total RNA from root and leaf samples was isolated using the guanidine thiocyanate extraction method with Trizol reagent (Vazyme, R401). RNA samples were treated with DNase I after the extraction to eliminate the potential trace contaminants of genomic DNA. For conducting qRT-PCR analysis, approximately 2 µg of total RNA from each sample was used to synthesize first-strand cDNA, and qRT-PCR assay was performed with ChamQ SYBR color qPCR Master Kit (Vazyme, Q411) on the LightCycler96 Systems (Roche). The relative transcript abundance of each target gene was standardized to the transcript level of a constitutive Actin gene. The primers used for qRT-PCR were listed in the Table S2.
Results
OsPT6 promoter confers PSI expression of GUS reporter gene in transgenic rice plants
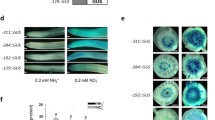
We have previously reported a rice phosphate transporter gene, OsPT6, that showed a strong up-regulation in response to Pi deficiency (Ai et al. 2009; Li et al. 2014). To explore in more detail the expression patterns of OsPT6, a 2860-bp promoter fragment of OsPT6 (P-2860) was fused to the GUS reporter gene and introduced into rice plants. Two-week-old seedlings of individual T2 transgenic lines were transferred to hydroponic culture treated with either high Pi (300 μM) or low Pi (15 μM) for 3 weeks. The GUS staining directed by the OsPT6 promoter was barely detectable in both blades and roots when sufficient Pi was supplied (Fig. 1), whereas under low Pi supply, strong GUS staining was observed throughout the primary and lateral roots except for some parts of the root tip. A strong GUS staining could also be observed in the blades of the rice plants supplied with low Pi. These results highlight that the 2,860-bp OsPT6 promoter fragment contains essential cis-elements necessary to confer Pi-deficiency responsive expression in rice.
Histochemical staining for the promoter activity of OsPT6 in response to Pi starvation using the GUS reporter gene. a–c GUS staining driven by the OsPT6 promoter in the root tips (a), lateral roots (b) and blades (c) of transgenic rice plants grown under low Pi (15 µM). (d–f) GUS staining in the root tips (d), lateral roots (e) and blades (f) of transgenic rice plants grown under high Pi (300 µM)
To screen the potential motifs responsible for the PSI response in OsPT6 promoter, we further performed motif analysis using the PLACE algorithm (http://www.dna.affrc.go.jp/PLACE/signalscan.html). Multiple regulatory motifs, including the well established Pi-starvation-responsive element, P1BS (GNATATNC), and some other elements, such as W-box (TTGACC) and PHO-Like, that have also been suggested to be associated with Pi signaling, were found to be present in the OsPT6 promoter (Table S1).
The PSI response of OsPT6 was regulated by at least two functional regions in its promoter
To gain further insights into the regulatory mechanism underlying the PSI response of OsPT6, a series of promoter truncations (Fig. 2a) fused to the GUS reporter gene were generated and introduced into transgenic rice plants. Seedlings of each transgenic line were treated with high Pi (300 μM Pi, HP) and low Pi (15 μM Pi, LP), respectively, for 3 weeks. As expected, none of the transgenic plants harboring the truncated promoter fragments showed an observable GUS staining in both blades and roots under the high Pi supply (Fig. 2b). A truncated 825 bp promoter fragment (P-825) upstream of the start codon ATG could drive the GUS expression highly in both the roots and blades of transgenic rice plants grown under low Pi. An detectable but much weaker GUS staining could be observed in the P-687 plants. A further deletion of the OsPT6 promoter down to -99 upstream of the ATG start codon (P-99) resulted in almost the complete absence of GUS staining in both roots and blades of all the transgenic plants (Fig. 2b). GUS enzymatic activity assay showed that the P-825 plants had comparable GUS activity with that in the plants harboring the full-length OsPT6 promoter (P-2860) (Fig. 3). By contrast, GUS enzymatic activity in the P-687 plants was decreased by nearly 75% compared with that in the P-2860 and P-825 transgenic plants. These results suggest the likely presence of cis-acting elements conferring the PSI response in both the − 825 to − 687 and − 687 to − 99 regions.
OsPT6 promoter deletion constructs and their activity in response to Pi starvation. a Different lengths of OsPT6 promoter fragment containing or not the P1BS and W-box motifs. The locations of P1BS and W-box motifs on OsPT6 promoter were labeled with triangle and ellipse, respectively. b GUS staining driven by different length of OsPT6 promoter fragment in root tips, lateral roots and blades of transgenic rice plants grown under low Pi (LP, 15 µM) and high Pi (HL, 300 µM) conditions
GUS enzymatic activity driven by different OsPT6 promoter fragments under low P and high P growth conditions. Seedlings of transgenic rice plants containing different OsPT6 promoter fragments were supplied with either low Pi (LP, 15 µM) or high Pi (HP, 300 µM) for 3 weeks, and then the roots and shoots were sampled separately for assaying the GUS enzymatic activity. Asterisks indicate significant differences in GUS enzymatic activity compared with the treatment of high P (two-tailed paired Student’s t test, p ≤ 0.05). Values are means of three biological replicates with SE. Asterisks indicate significant differences. *p < 0.05; ***p < 0.001
P1BS is a key cis-acting element mediating OsPT6 responsive to Pi starvation
P1BS is a conserved Pi-starvation responsive element that is abundantly present in the promoters of many PSI genes (Rubio et al. 2001; Bustos et al. 2010; Li et al. 2014). There is only one copy of the P1BS motif (− 107 bp upstream of the start codon) in the OsPT6 promoter (Table S1, Fig. 2a). In our previous study we have proven that OsPT6 prompter could drive a purple gene (Pr) isolated from cauliflower to express strongly in transgenic tobacco plants under low Pi conditions (Li et al. 2014), indicating that the regulatory components responsible for modulating the PSI response of OsPT6 is highly conserved across dicot and monocot species. To determine whether the P1BS is essential for the PSI response of OsPT6, we deleted of the P1BS motif from the P-2860 fragment, and the modified promoter fragment, named △P1BS, was fused to the GUS reporter gene. The resulting construct was then transformed into the tobacco plants, and the transgenic tobacco harboring the P-2860-GUS vector was used as a positive control. The GUS induction rate which indicates the ratio of GUS enzymatic activity in the corresponding plants grown under low P and high P was assayed. Unlike the P-2860 plants having a high GUS induction in response to low Pi, GUS activity in the △P1BS plants supplied with low Pi was comparable with that in the high-Pi-treated plants (Fig. 4b), indicating that absence of P1BS in OsPT6 promoter caused a complete loss of the ability to induce the GUS expression under low Pi.
Functional analysis of the P1BS motif involved in the PSI activation of OsPT6. a Schematic illustration of the P1BS deletion construct △P1BS. The locations of P1BS and W-box motifs on OsPT6 promoter were labeled with ellipse and triangle, respectively. b GUS induction rate, which indicates the ratio of GUS enzymatic activity in the corresponding plants grown under low P and high P, was assayed in the roots of P-2860 and △P1BS transgenic tobacco plants. c Schematic illustration of the construct of 4X P1BS repeats fused to the minimal 35S promoter (mini35S Pro) driving the GUS reporter gene. d The relative GUS transcription level was assayed by qRT-PCR in the transgenic tobacco plants harboring the 4X P1BS-GUS construct under low P and high P conditions. Asterisks indicate significant differences in GUS induction rate compared with the group of control (two-tailed paired Student’s t test, ***p < 0.001). Values are means ± SE(n = 3). Error bars denote SE
To further verify the capacity of the P1BS motif in response to Pi starvation, we designed an artificial construct (4XP1BS-GUS), which consists of four tandem repeats of the P1BS motif from the OsPT6 promoter fusing to a 35S minimal promoter (Fig. 4c), and the resulting construct was transformed into Nicotiana benthamiana leaves via Agrobacterium-mediated transient transformation. Four-week-old seedlings were transferred to hydroponic culture treated with either high Pi (1 mM) or low Pi (15 μM) for 7 days. The relative transcripts of GUS reporter gene was assayed by real-time quantitative PCR. As shown in Fig. 4d, Pi-depleted plants expressing the 4XP1BS::GUS chimeric gene showed a much higher GUS induction than those grown under high Pi conditions. These results strongly suggest an essential role of the P1BS element in maintaining the PSI response of OsPT6.
The downstream W-box is also involved in the regulation of PSI response of OsPT6
W-box motif has also be documented to be associated with the regulation of genes involved in Pi signaling (Wang et al. 2014a, b). As two copies of W-box (− 704 and −2281 upstream of the start codon, respectively) are present in the OsPT6 promoter, it inspires us to verify whether the two W-box motifs are also involved in the regulation of OsPT6 responsive to Pi starvation. To this end, we performed targeted deletion of one or both of them in the P-2860 promoter fragment. We also generated a construct with deletions for both the two W-box and P1BS motifs (Fig. 5a).These resulting constructs, named △w1, △w2, △w1w2, and △w1w2-P, respectively, were transformed into the tobacco plants. Compared to P-2860 plants, deletion of the upstream W-box motif (W1) did not showed a significant difference in the GUS induction rate in the △w1 plants (Fig. 5b). However, deletion of the downstream W-box (W2) or simultaneous deletion of the w1 and w2 resulted in a significant reduction in the GUS induction rate in these corresponding plants (Fig. 5b). Moreover, the △w2 plants were shown to have a comparable level of GUS induction rate with those △w1w2 plants. These results indicate that W2, but not W1, is involved in the regulation of PSI response of OsPT6. The △w1w2-P plants showed a decrease by approximately 60% in GUS induction rate compared with the △w1w2 plants. However, no significant difference in GUS induction rate was observed between the △w1w2-P plants and the △P1BS plants, suggesting that the W-box-mediated regulation of PSI response of OsPT6 might be highly dependent on the P1BS motif.
Functional analysis of the two W-box motifs involved in the PSI activation of OsPT6. a Schematic illustration of the different constructs containing one or both deletions of W-box motifs. The locations of P1BS and W-box motifs on OsPT6 promoter were labeled with ellipse and triangle, respectively. b GUS induction rate was assayed in the roots of transgenic tobacco plants harboring different W-box deletion constructs. Asterisks indicate significant differences in GUS enzymatic activity compared with the P-2860 promoter fragment (two-tailed paired Student’s t test, p ≤ 0.05). Values are means of three biological replicates with SE. Asterisks indicate significant differences. **p < 0.01; ***p < 0.001
Discussion
Low availability of P is often a major limiting factor to crop yields in most nutrient-poor soils. Transcriptional up-regulation of genes involved in Pi sensing, transport and redistribution upon Pi starvation is one of the most efficient strategies evolved by plants to adapt to limited Pi supplies (López-Arredondo et al. 2014; Ha et al. 2014). Identifying regulatory components, including cis-acting elements and their binding transcription factors, involved in the regulation of the expression of PSI genes is an important step towards better understanding the Pi signaling network in certain plant species (Wu et al. 2013; Liang et al. 2014).
Promoter deletion assay is a common and effective approach to identify cis-acting elements responsible for tissue-specific or development-dependent expression patterns. In this study, promoter truncation analysis on OsPT6, a rice Pi transporter gene strongly responsive to Pi starvation (Fig. 1), revealed the presence of at least two regulatory regions (− 825 to − 687 and − 687 to − 99) that are required for the regulation of PSI response of OsPT6 (Figs. 2b, 3). Promoter cis-element analysis using the PLACE algorithm led to the identification of several regulatory motifs, such as P1BS and W-box, that are probably associated with Pi signaling in the OsPT6 promoter (Table S1, Fig. 2a). As P1BS element has been proven to be a key regulator of several Pi transporter genes responsive to Pi starvation and even mycorrhizal colonization, and there is only one copy of the P1BS element in the OsPT6 promoter, it is tempting to speculate that this P1BS element might be the suitable candidate responsible for the up-regulation of OsPT6 in response to Pi starvation. The function of P1BS in PSI expression was further evidenced by its deletion from the full-length OsPT6 promoter (P-2860). Targeted deletion of this P1BS motif resulted in almost complete absence of GUS induction by Pi starvation in △P1BS plants (Fig. 4b), indicating that P1BS is essential for the activation of PSI response of OsPT6. A four-tandem repeat of the P1BS motif from the OsPT6 promoter fused to the minimal 35S promoter that was able to promote GUS induction rate (Fig. 4d) provides further indirect support for its capacity in inducing the Pi starvation-responsive expression in rice.
The W-box motif is another regulatory element that has been experimentally validated to mediate Pi starvation-responsive expression by interaction with the WRKY transcription factors. Several WRKY proteins have been shown to be up-regulated upon Pi starvation and act as positive regulators for Pi transporter genes, such as Arabidopsis AtPT1 (Wang et al. 2014a, b). However, previous studies have also proposed a negative role of the interactions between the W-box element and WRKY proteins in regulating Pi-related genes, such as PHO1. In Arabidopsis, WRKY6 and WRKY42 suppress PHO1 expression through binding to the W-box motifs in its promoter (Chen et al. 2009; Su et al. 2015). These findings suggest that the W-box/WRKY assembly could serve as both positive and negative regulators of a certain of Pi physiological processes. In this study two copies of W-box motifs were found to be existent in the OsPT6 promoter (Table S1). Target deletion of the upstream W-box motif (W1) in OsPT6 promoter did not significantly affect the GUS induction rate in △w1 plants compared with that in the P-2860 plants (Fig. 5). This gives a strong hint that the W1 motif that is far away from the start codon might have no substantial role in the up-regulation of OsPT6 responsive to Pi starvation. This hypothesis could gain well support from the fact that the P-825 truncation that is absence of the W1 had a comparable capacity with the P-2860 fragment in driving the GUS expression in response to Pi starvation (Fig. 3). A positive role of the downstream W-box motif (W2) in promoting OsPT6 expression under Pi starvation could be deduced from that deletion of W2 or both of the two W-box motifs resulted in a remarkable reduction in GUS induction in △w2 or △w1w2 plants compared with that in the P-2860 plants. The absence of regulatory capacity with respect to the W1 motif might be due to its inefficient flanking sequence that hinders the binding of W1 to the potential WRKY proteins. Such a case could also be observed from a previous study reporting that WRKY6 represses PHO1 expression to balance Pi homeostasis in Arabidopsis through its binding to two of the six W-box motifs in the PHO1 promoter (Chen et al. 2009).
Conclusion
In the present work, through a series of promoter truncation/deletion analysis, we identified two conserved cis-acting motifs, the P1BS motif and the downstream W-box motif, that are involved in the regulation of PSI expression of OsPT6. We proposed that both of the two motifs act as positive regulators in directing the expression of OsPT6 in response to Pi starvation, and the W-box-mediated regulation of PSI response of OsPT6 might be highly dependent on the P1BS motif. Given the complexity of the Pi signaling regulation in plants, we could not rule out the possibility that the PSI response of OsPT6 could also be co-regulated by other cis-acting elements in its promoter. As most Pht1 genes in Arabidopsis and rice contain P1BS and W-box motifs in their promoters, our findings obtained from this study may provide some clues for further studying the regulatory mechanisms of other Pht1 genes in response to Pi starvation.
References
Ai PH, Sun SB, Zhao JN, Fan XR, Xin WJ, Guo Q, Ling Y, Shen QR, Wu P, Miller AJ, Xu GH (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57(5):798–809
Bustos R, Gabriel C, Francisco L, Isabel PM, Vicente R, Julian P-P, Roberto S, Antonio L, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6(9):110–117
Chen YF, Li LQ, Xu Q, Kong YH, Wu WH (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21(11):3554–3566
Clarkson DT, Hanson JB (2003) The mineral nutrition of higher plants. Ann Rev Plant Physiol 31(1):239–298
Dai XY, Wang YY, Zhang WH (2016) OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice[J]. J Exp Bot 3:947–960
Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143(4):1789–1801
Ferrol N, Azcón-Aguilar C, Pérez-Tienda J (2019) Review: Arbuscular mycorrhizas as key players in sustainable plant phosphorus acquisition: An overview on the mechanisms involved[J]. Plant Sci 280:441–447
Fraley RT, Horsch RB (1983) In vitro plant transformation systems using liposomes and bacterial co-cultivation. Genetic Engineering of Plants, pp. 177–179, Plenum Pub. Co., New York.
Gu M, Zhang J, Li HH, Meng DQ, Li R, Dai XL, Wang SC, Liu W, Qu HY, Xu GH (2017) Maintenance of phosphate homeostasis and root development are coordinately regulated by MYB1, an R2R3-type MYB transcription factor in rice. J Exp Bot 13:3603–3615
Guo MN, Ruan WY, Li CY, Huang FL, Zeng M, Liu YY, Yu YN, Ding XM, Wu YR, Wu ZC, Mao CZ, Yi KK, Wu P, Mo XR (2015) Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol 168(4):1762–76
Ha S, Tran LS (2014) Understanding plant responses to phosphorus starvation for improvement of plant tolerance to phosphorus deficiency by biotechnological approaches. Crit Rev Biotechnol 34(1):16–30
Hernando N, Wagner CA (2018) Mechanisms and regulation of intestinal phosphate absorption. Compr Physiol 8(3):1065–1090
Jia HF, Ren HY, Gu M, Zhao JN, Xu GH (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156(3):1164–1175
Karthikeyan AS, Ballachanda DN, Raghothama KG (2009) Promoter deletion analysis elucidates the role of cis elements and 5’UTR intron in spatiotemporal regulation of AtPht1;4 expression in Arabidopsis. Physiol Plantarum 136(1):10–18
Lambers H, Martinoia E, Renton M (2015) Plant adaptations to severely phosphorus-impoverished soils. Curr Opin Plant Biol 25:23–31
Lapis-Gaza HR, Jost R, Finnegan PM (2014) Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol 14:334
Li, YT, Gu M, Zhang X, Zhang J, Fan HM, Li PP, Li ZF, Xu GH (2014) Engineering a sensitive visual-tracking reporter system for real-time monitoring phosphorus deficiency in tobacco. Plant biotechnol J
Li YT, Zhang J, Zhang X, Fan HM, Gu M, Qu HY, Xu GH (2015) Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci 230:23–32
Liang C, Wang J, Zhao J, Tian J, Liao H (2014) Control of phosphate homeostasis through gene regulation in crops. Curr Opin Plant Biol 21:59–66
Lin WY, Lin SI, Chiou TJ (2009) Molecular regulators of phosphate homeostasis in plants. J Exp Bot
Liu F, Wang ZY, Ren HY, Shen CJ, Li Y, Ling HQ, Wu CY, Lian XM, Wu P (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62(3):508–517
López-Arredondo DL, Leyva-González MA, Antonin M, González-Morales SI, López-Bucio J, Herrera-Estrella L (2014) Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol 65(1):95–123
Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55(5):727–741
Muchhal USU, Pardo JMJ, Raghothama KGK (1996) Phosphate transporter from higher plant Arabidopsis thaliana. Proc Natl Acad Sci 93(19):10519–10523
Nagarajan VK, Jain A, Poling MD, Lewis AJ, Raghothama KG, Smith AP (2011) Arabidopsis Pht1;5 Mobilizes Phosphate between Source and Sink Organs and Influences the Interaction between Phosphate Homeostasis and Ethylene Signaling. Plant Physiol 156(3):1149–1163
Nussaume L, Kanno S, Javot H, Marin E, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2(83):83
Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50:665–693
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15(16):2122–2133
Sawers RJH, Svane SF, Quan C, Grønlund M (2017) Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root-external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol 214(2):632–643
Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant Journal 39(4):629–642
Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, Chen YF (2015) WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in arabidopsis. Plant Physiol 167(4):1579–1591
Sun SB, Gu M, Cao Y, Huang XP, Zhang X, Ai PH, Zhao JN, Fan XR, Xu GH (2012) A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol 159(4):1571–1581
Upadhyaya NM, Surin B, Ramm K, Gaudron J, Schünmann PHD, Taylor W, Waterhouse PM, Wang MB (2000) Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers[J]. Austral J Plant Physiol 27:201–210
Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157(3):423–447
Wang H, Xu Q, Kong YH, Chen Y, Duan JY, Wu WH, Chen YF (2014a) Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1; 1 Expression In Response To Phosphate Starvation1 W OPEN. Plant Physiol 164(4):2020–2029
Wang XF, Wang YF, Piñeros MA, Wang ZY, Wang WX, Li CY, Wu ZC, Kochian LV, Wu P (2014b) Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant, Cell Environ 37(5):1159–1170
Wang XF, Deng MJ, Xu JM, Zhu XL, Mao CZ (2018) Molecular mechanisms of phosphate transport and signaling in higher plants. Semin Cell Dev Biol 74:114–122
Wu P, Shou HX, Xu GH, Lian XM (2013) Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr Opin Plant Biol 16(2):205–212
Ye Y, Yuan J, Chang XJ, Yang M, Zhang LJ, Lu K, Lian XM, Hatem R (2015) The Phosphate Transporter Gene OsPht1;4 Is Involved in Phosphate Homeostasis in Rice. PloS one 10(5):e0126186
Yi KK, Wu ZC, Zhou J, Du LM, Guo LB, Wu YR, Wu P (2005) OsPTF1, a Novel Transcription Factor Involved in Tolerance to Phosphate Starvation in Rice. Plant Physiol 138(4):2087–2096
Zhou J, Jiao FC, Wu ZC, Li YY, Wang XM, He XW, Zhong WQ, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146(4):1673–1686
Acknowledgements
This study is funded by the grants from National Natural Science Foundation of China (Grant No. 31501817) and the agricultural science and technology innovation program (ASTIP-TRIC01-2020).
Author information
Authors and Affiliations
Contributions
Designing the research, YL, and GX, Performing the experiments, QZ, ZL, and JC, Analyzing the data, HJ, and PA, Writing the manuscript, YL, Supervision, AC, and GX.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by Dawei Xue.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, Q., Luo, Z., Chen, J. et al. Characterization of two cis-acting elements, P1BS and W-box, in the regulation of OsPT6 responsive to phosphors deficiency. Plant Growth Regul 93, 303–310 (2021). https://doi.org/10.1007/s10725-020-00688-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-020-00688-z