Identification of Genome Sequences of Polyphosphate-Accumulating Organisms by Machine Learning
- State Key Laboratory of Urban Water Resource and Environment, School of Environment, Harbin Institute of Technology, Harbin, China
In the field of sewage treatment, the identification of polyphosphate-accumulating organisms (PAOs) usually relies on biological experiments. However, biological experiments are not only complicated and time-consuming, but also costly. In recent years, machine learning has been widely used in many fields, but it is seldom used in the water treatment. The present work presented a high accuracy support vector machine (SVM) algorithm to realize the rapid identification and prediction of PAOs. We obtained 6,318 genome sequences of microorganisms from the publicly available microbial genome database for comparative analysis (MBGD). Minimap2 was used to compare the genomes of the obtained microorganisms in pairs, and read the overlap. The SVM model was established using the similarity of the genome sequences. In this SVM model, the average accuracy is 0.9628 ± 0.019 with 10-fold cross-validation. By predicting 2,652 microorganisms, 22 potential PAOs were obtained. Through the analysis of the predicted potential PAOs, most of them could be indirectly verified their phosphorus removal characteristics from previous reports. The SVM model we built shows high prediction accuracy and good stability.
Introduction
Phosphorus (P) is one of the key element controlling the normal functioning of many organisms in the ecosystem (Zeng et al., 2004). In aqueous environment, phosphorus compounds could be hydrolyzed to orthophosphate, the only form of phosphorus which could be utilized by aquatic organisms (Correll, 1998). However, presence of excess phosphate into the water bodies, carried by domestic and industrial wastewater, may trigger eutrophication, which lead to algae bloom, oxygen depletion in water, and biological organism death (Smith et al., 1999; Ju et al., 2016). Study shows that about 260,000 metric tons of P were discharged from wastewater treatment facilities every year in the US (Litke, 1999; Loganathan et al., 2014). With the significant increase of phosphorus levels in the water environment, people are concerned about the deterioration of water quality and overall ecological balance (Stoddard et al., 2016).
Among of all methodologies, biological phosphorus removal (BPR), as the most economical strategy was widely used in wastewater treatment plants (WWTP) (Huang et al., 2020). Previous studies suggested that polyphosphate-accumulating organisms (PAOs) are able to remove P removal from BPR system (Cai et al., 2019; Gao et al., 2019; Wang et al., 2019). It is mainly because of the special metabolism of PAOs under alternative anaerobic and aerobic reaction. Under the anaerobic reaction, the polyP stored in PAOs are degraded to orthoP and releases into the liquid with the orthoP adsorbed by EPS. Under the aerobic reaction, a part of orthoP passes through EPS and stores P as polyP inside PAOs, and the other part can be adsorbed by the surrounding EPS matrix (Liu et al., 2020). One of such study reported that over 90% P was removed under aerobic environment in a sequencing batch reactor by PAOs. Therefore, it is feasible to increase the content of PAOs in the activated sludge to improve the P removal efficiency. The pre-requisite for increasing the content of PAOs is to be able to effectively identify PAOs. Recently, some work has been reported in the identification of PAOs, just like Dechloromonas (Kong et al., 2007; Günther et al., 2009), Candidatus accumulimonas spp. (Nguyen et al., 2012), Microlunatus spp. (Kawaharasaki et al., 1999; Beer et al., 2004), Aeromonas (Wang et al., 2009), Tetrasphaera spp. (Kong et al., 2005; Nguyen et al., 2011), Pseudomonas (Shi and Lee, 2006), Klebsiella (Sun et al., 2016), Rhodopseudomonas (Tsuneda et al., 2005), and so on. However, due to the variety of PAOs, although more than 7,000 species have been identified, there are still many strains of PAOs that have not been found. At present, most of the existing researches on PAOs were based on gene-level analysis. The species and the relative abundance of PAOs of biological phosphorus removal system could be investigated by high-throughput sequencing technology (Lin et al., 2019; Salehi et al., 2019; Xu et al., 2019). However, it is cumbersome and time-consuming to sieve and identify the PAOs by conventional experimental methods. Therefore, it is particularly important to find a fast and effective method to identify the PAOs.
Recently, algorithms have been widely used in medicine, geology, hydrology and so on, due to its convenience, quickness and accurate prediction (Fijani et al., 2018; Pham et al., 2018; Peng and Zhao, 2020; Zhao et al., 2020a,b,c,d). Pang et al. (2019) developed a fluctuant influent responsive Q-learning based BPR optimizing control method. An efficient approach based on bi-sensitivity analysis and genetic algorithm for calibration of activated sludge models was proposed by Chen et al. (2015). However, the algorithm for the identification of PAOs has not been reported yet. Identification of PAOs, which is described as a classification problem, can be achieved using supervised classification algorithms. The support vector machine (SVM) is an effective and intelligent machine learning method which can solve classification problems and non-linear function estimation problems in the fields of pattern recognition and machine learning (Vapnik, 1995; Chen et al., 2008; Cheng and Jhan, 2012). In the context of intelligent classification, SVM combined with feature extraction method has been successfully employed in the field of pattern recognition (Ballabio and Sterlacchini, 2012; Chen et al., 2017; Shi et al., 2017; Fijani et al., 2018; Fu et al., 2019). Besides, under the same detection performance conditions, SVM require less prior knowledge than other methods, and the training time can be greatly shortened, which provides favorable conditions for the identification of genome sequences. To the best of our knowledge, there are few studies till now applying SVM to distinguish and identify PAOs in the complex biological activated sludge.
In this study, a SVM algorithm was proposed to realize the rapid identification of PAOs in activated sludge system. The genome sequences alignment of thousands of bacterial groups were used to find out the characteristic genome of PAOs. The main objective of this study was to establish a new model through the SVM algorithm, which can use the features of the genome sequences to achieve large-scale and high-precision identification of PAOs, with an accuracy of over 90%. This development illustrated a novel and efficient strategy to accelerate the screening and identification of microorganism.
Materials and Methods
Method Overview
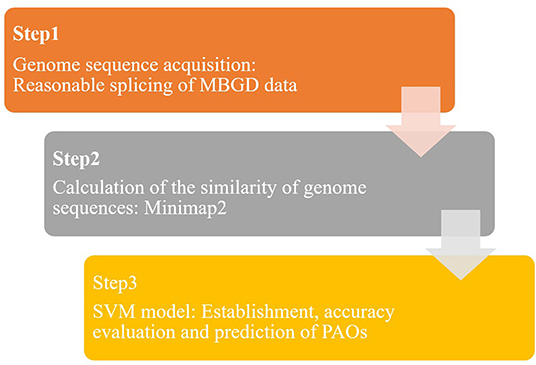
In this study, a predictive model of PAOs was established based on the SVM algorithm, to realize rapid and accurate identification of PAOs. The SVM-based identification method of PAOs was divided into three steps (Figure 1): genome sequences collection, pairwise alignment for genome sequences and the identification of PAOs based on SVM.
Step 1: The genome sequences of microorganisms were extracted from the publicly available microbial genome database for comparative analysis (MBGD). New long-chain genome sequences were obtained by splicing the genome sequences in MBGD database to serve as training databases.
Step 2: Data processing was conducted to get a series of s the similarity between genome sequences, based on the database formed from MBGD (Step 1). Minimap2 was used to compare the obtained genome sequences in pairs, and read the overlap. By calculating the overlap, the similarity between genome sequences was achieved.
Step 3: The SVM model based on the prediction of PAOs was established. Through feature selection of the above similarity, we got a matrix of 1*6,318. The accuracy of the SVM model is verified by grouping the microorganisms reasonably. In addition, over 2,000 species of bacteria were identified based on this SVM model to predict the potential PAOs.
Genome Sequences Collection
For this study, all the genome sequence datasets were retrieved from the publicly available microbial genome database for comparative analysis (MBGD, available at http://mbgd.genome.ad.jp/). As its unique feature, MBGD allows users to perform orthology analysis among any specified set of organisms (Uchiyama et al., 2013). It is this flexibility that makes MBGD adapt to a variety of microbial genomic study. Due to the huge diversity of microorganisms, a total of 6,318 representative microbial genome sequences including 5,861 bacteria, 254 archaea and 203 eukaryota stored in the MBGD were used for analysis. However, the gene sequence of each microorganism provided by MBGD is divided into several thousand groups that cannot be directly compared. Therefore, these genome sequences were spliced to reconstitute a new genome sequence for similarity comparison.
Similarity Comparison of Genome Sequences
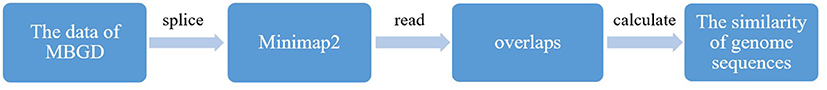
By splicing the genome sequences of microorganisms provided by the MBGD, each microbe has obtained a long-chain gene. In this study, we used minimap2 to rapidly obtain all-to-all read overlaps (Figure 2). Minimap2 was a first DNA and RNA-seq aligner specifically designed for long sequence alignment (Li, 2018). In terms of long sequence alignment, minimap2 achieved approximate mapping 50 times faster than BWA-MEM (Li, 2016). Minimap2 is based on identifying reads that share many co-linear minimizers (Roberts et al., 2004). Minimap2 uses indexing and seeding algorithms similar to minimap (Li, 2016), and furthers the predecessor with more accurate chaining, the ability to produce base-level alignment and the support of spliced alignment. We used minimap2 to compare the genomes of the obtained microorganisms in pairs, and read the overlap. By calculating the overlaps, the similarity between the microbial genome sequences were available.
Support Vector Machine (SVM)
The support vector machine (SVM) is one of the most successful machine learning algorithm in the prediction field developed by Vapnik (1995). It has been widely applied in bioinformatics, computational biology and environmental studies (Chen et al., 2012; Liu and Lu, 2014; Ding et al., 2016; Pan et al., 2018). The SVM model is used to find the optimal solution separating two classes which adopts the theory of structural risk minimization instead of empirical risk minimization to reduce the over-fitting problem. Therefore, the SVM algorithm is very effective for the identification and prediction of PAOs. In addition, unlike traditional machine learning, SVM can convert the complex non-linear problem input data to a higher-dimensional space where a hyperplane is constructed by introducing kernel function (Vapnik and Vapnik, 1998; Carrier et al., 2013; Ch et al., 2013; He et al., 2014). The main idea of SVM is to create a line or a hyperplane as the decision surface that maximizes the margin between two classes. The hyperplane function can be defined as follows:
where w represents the weight vector, b is the bias parameter, and x indicates the input vector in the sample space. Taking binary classification problem as an example, to correctly classify samples, all samples are required to meet the following constraints:
However, in practice, some abnormal points that cannot be linearly separated often exist. In order to solve this problem, a slack variable ξi and penalty factor C are added. The optimization function and constraints are as follows:
For simplifying the calculation, the above equation can be transformed into solving the saddle point of the Lagrange equation by using the Lagrange multiplier. By applying the duality theorem, the final optimization problem is presented as:
where λi is Lagrange multiplier.
This problem can be solved by using quadratic programming. The final linear discriminant function that is used for the classification of new data can be achieved as follows:
However, after mapping the original space into the higher dimensional feature space, different inner product kernel functions will form different algorithms. As we know, four types of kernel functions are available in SVM including polynomial kernel (PL), sigmoid kernel (SIG), radial basis function (RBF) and linear kernel (LN). Besides, the prediction accuracy would be also enhanced with appropriate kernel function. In the previous stage, we have performed a pairwise comparison of the gene sequences of the 6,318 strains in MBGD. By calculating the similarity, we get a matrix of 1*6318. Since the feature dimension we got is already very high, there is no need to perform higher-dimensional mapping by other kernel functions. Therefore, in this method, we employed the LN function to train our predictive models, which is defined as:
The kernel function is used to transform data into two classes consisting of unknown bacteria and PAOs {0, 1}.
The optimal classification function can be constructed as follows:
Results
Data Pre-processing
Experiments adopted an MBGD data which combined the Reference Sequences (RefSeq) database of National Center for Biotechnology Information (NCBI), the original GenBank entry, and the Gene Trek in Prokaryote Space (GTPS) provided by DNA Data Bank of Japan (DDBJ). The genome sequences of 6,318 microorganisms were provided by MBGD. However, the genome sequence of each microorganism provided by MBGD is divided into several thousand groups that cannot be directly compared. Therefore, we made a reasonable splicing of the microbial genome sequences, so that each microbial obtained a long chain of genome sequence. Feature selection was used before SVM algorithm for choosing a subset of the features that is clean of redundancies. Moreover, it is necessary to select highly correlated features over other features to improve the prediction accuracy in the SVM model. After obtaining the microbial genome sequences, minimap2 which is a general-purpose alignment program to map DNA or long mRNA sequences against a large reference database was used to compare the genome sequences, and read the overlap. By calculating the overlaps, the similarity between the microbial genome sequences were available. We obtained a matrix of 1*6318 to complete the feature selection.
SVM Experiments
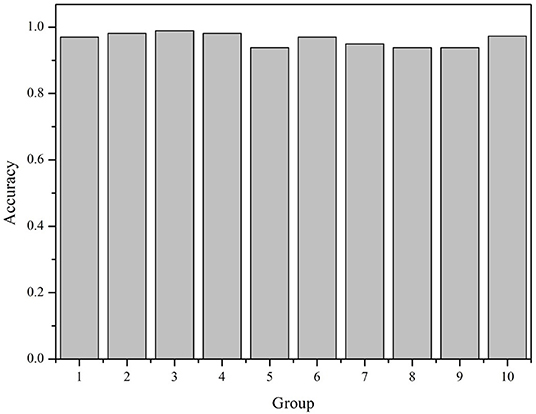
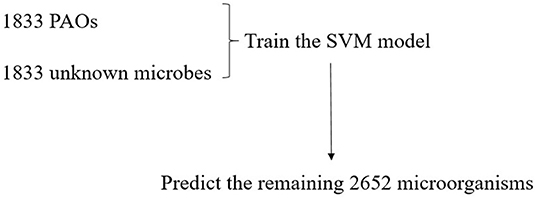
All the 6,318 microbial genome sequences were provided by MBGD, including 1,833 known PAOs, such as Dechloromonas, Pseudomonas, Rhodopseudomonas, and Klebsiella. The remaining 4,485 microbes have not been proved whether they are PAOs. All the 1,833 known PAOs and the remaining 4,485 microbes were selected to form the sample database of the SVM model. All the data were classified into [0, 1], 0 for the microorganisms that are uncertain whether they are PAOs, and one for the PAOs. Subsequently, the features obtained were trained by the SVM classifier, which were then evaluated with 10-fold cross-validation to test the accuracy of the algorithm (Figure 3). 10-fold cross-validation is a common test method which is to divide the sample database into ten parts, and takes turns using nine parts as training data and one part as test data for testing. The corresponding accuracy rate can be obtained in each experiment, and the average accuracy of the 10 results can be used to evaluate the accuracy of the algorithm. In the SVM model, the 1,833 known PAOs and the remaining 4,485 microbes were divided into 10 parts with 183~184 PAOs and 448~449 unknown microbes randomly. In order to improve the accuracy of cross-validation, the above 10 parts of data were divided into 10 groups randomly. Each group contained 18~19 PAOs and 44~45 unknown microbes. For the 10 groups of data in each part, we randomly selected nine groups of data to train the SVM model, and the remaining one group of data for testing. In other words, of all the data, 90% were used for training and 10% were used for testing. The Figure 4 illustrated the satisfactory prediction accuracy of the SVM models in the testing stage. The test accuracy of the SVM model for 10 parts of data are: 0.9700, 0.9810, 0.9889, 0.9810, 0.9383, 0.9699, 0.9493, 0.9382, 0.9382, and 0.9731. The average accuracy provided by the SVM model is 0.9628±0.019. Therefore, the SVM model shows high prediction accuracy and good stability, and can be used for the prediction of PAOs in the future.
Discussion
Due to the high accuracy and good stability of the SVM model, it can be used to predict PAOs. In this experiment, we selected 1,833 of all known PAOs, and randomly selected 1,833 from the remaining microorganisms as the negative samples to test the model. Subsequently, we made predictions on the remaining 2,652 microorganisms and identified potential PAOs that may not be discovered (Figure 5).
It can be seen from the prediction results that we have predicted 22 potential PAOs, namely: Anoxybacillus flavithermus WK1, Arcanobacterium haemolyticum DSM 20,595, Anoxybacillus sp. B7M1 b7m1, Anoxybacillus sp. B2M1 b2m1, Arthrobacter phenanthrenivorans Sphe 3, Amphilbacillus xylanus NBRC 15112, Brevibacillus brevis NBRC NBRC 100599, Corallococcus coralloides DSM 2259, Arthrospira sp. PCC 8005, Salmonella enterica subsp. Enterica serovar Choleraesuis SCSA50, Campylobacter jejuni subsp. jejuni RM3196, Pannonibacter phragmitetus 31801, Helicobacter pylori ML1, Shigella flexneri 2a 981, Lactobacillus plantarum JBE245, Sulcia muelleri PUNC, Jannaschia sp. CCS1, Kinetoplastibacterium oncopeltii TCC290E, Thiomonas sp. 3As, Teredinibacter turnerae T7901, Wolbachia endosymbiont TRS, Wolbachia endosymbiont of Drosophila simulans wHa. All the above 22 kinds of microorganisms are potential PAOs which have not been reported, and their characteristics are analyzed as follows.
A. flavithermus WK1 was isolated from the waste water drain at the Wairakei geothermal power station in New Zealand (Mountain et al., 2003). It is a member of the family Bacillaceae. A. flavithermus grow only at ~30~72°C (Heinen et al., 1982), which are always found in high-temperature habitats such as geothermal hot springs, the manufacture of milk powder, and manure (Heinen et al., 1982; Pikuta et al., 2000; Clerck et al., 2004; Rueckert et al., 2005). So, it is a typical thermophilic bacteria strain. Activated sludge process is generally conducted under the conditions of 12 °C~25 °C, which is not conducive to the growth of A. flavithermus WK1. Therefore, it has not been reported that A. flavithermus WK1 is PAOs. A. haemolyticum DSM 20595, a Gram-positive bacterium, can cause a wide range of diseases in humans, just as wound infections and pharyngitis (Banck and Nyman, 1986; Miller et al., 1986; Mackenzie et al., 1995; Linder, 1997). A. haemolyticum was previously in the Corynebacterium genus, but it is classified as a member of Actinobacteria now (Collins et al., 1982). It has been reported that most of the Actinobacteria may be PAOs, so we conclude that A. haemolyticum DSM 20595 is most likely to be a potential PAOs. Both Anoxybacillus sp. B7M1 b7m1 and Anoxybacillus sp. B2M1 b2m1 belong to Anoxybacillus which is closely related to the genus Geobacillus and the member of the family Bacillaceae (Pikuta et al., 2000). All of them are Gram-positive bacterium and thermophiles with optimum growth temperature around 55 °Cwhich are similar to the A. flavithermus WK1 (Goh et al., 2013). Besides, members of Anoxybacillus have been proved great potential in environmental applications (Zitomer et al., 2007; Ghaffari et al., 2011). Therefore, Anoxybacillus sp. B7M1 b7m1 and Anoxybacillus sp. B2M1 b2m1 are likely to have a special capture effect on phosphate. Amphibacillus xylanus strains were previously isolated from an alkaline compost. The genome of A. xylanus NBRC 15112 (GenBank: AP012050.1; Taxonomy ID: 698758) was released in 2013, and the type strain was confirmed to utilize xylan from oat spelt and larchwood, and to grow between pH 8 and pH 10 (Niimura et al., 1990). Amphibacillus is a member of Bacillus which is considered as PAOs. Therefore, A. xylanus NBRC 15112 is also likely to be a potential PAOs. A. phenanthrenivorans Sphe 3 is a Gram-positive, aerobic, novel type strain of the genus Arthrobacter belonging to Actinobacteria which was isolated from a creosote-contaminated soil in Epirus, Greece (Kallimanis et al., 2007). It can grow on phenanthrene as the sole source of carbon and energy with a suitable temperature of 30 °C~37 °C and pH of 7.0~7.5 (Kallimanis et al., 2011). Just like A. haemolyticum DSM 20595, due to belong to Actinobacteria, it is most likely an undiscovered PAOs. B. brevis NBRC NBRC 100599 is a Gram-positive and spore-forming bacterium which has a broad-spectrum antimicrobial activity by secreting some functional metabolites (Song et al., 2012; Pawlowski et al., 2016). Besides, some report indicated that B. brevis might be an effective strain for the degradation of polycyclic aromatic hydrocarbons (Zhu et al., 2019). However, whether it also has effect of P removal is not yet known. A potentially novel energy taxis cluster was found in C. coralloides DSM 2259 that has been proved to be the same as Actinobacteria. Amongst 34 sequenced myxobacterial genomes, C. coralloides is the only species that encodes such a CSS cluster (Huntley et al., 2012). Maybe this cluster of genes gives it the characteristics of PAOs. P. phragmitetus 31801 was isolated from the blood sample of a patient with liver abscess (Wang et al., 2017). Currently, the studies on P. phragmitetus mainly focused on its bioremediation potentials including reduction of heavy metal chromium and detoxification of polycyclic aromatic compounds (PAHs) under extreme conditions (Borsodi et al., 2005; Xu et al., 2011; Shi et al., 2012; Wang et al., 2013). Jannaschia sp. strain CCS1is an ecologically relevant marine proteobacterium found in coastal and open surface waters (Bakolitsa et al., 2010). However, whether they can remove phosphate is not yet known. Arthrospira sp. PCC 8005, better known as spirulina, provides exceptional nutritional value which was selected by the European Space Agency (ESA) for its nutritive value and oxygenic properties in the Micro-Ecological Life Support System Alternative (MELiSSA) life support system. It is a photosynthetic prokaryote that plays a crucial role in the Earth's nitrogen and phosphorus cycles. Due to the unique photosynthesis, it has a certain absorption effect on phosphorus (Janssen et al., 2010; Deschoenmaeker et al., 2014). S. enterica subsp. Enterica serovar Choleraesuis SCSA50, C. jejuni subsp. jejuni RM3196 and S. flexneri 2a 981 can cause severe invasive disease in humans just like food-borne bacterial gastroenteritis and diarrheal disease (Hughes and Cornblath, 2005; Peng et al., 2009; Senior, 2009; Parker et al., 2015). The infection of these pathogens are caused by the consumption of contaminated food or drink in humans and animals. Similarly, H. pylori ML1 is associated with chronic active type B gastritis and peptic ulcer diseases (Chaun, 2001). These pathogens are not recommended for use in the field of water treatment, so whether they have the effect of removing phosphate is not yet known. L. plantarum JBE245 is a member of L. plantarum which is isolated from malolactic fermentation of apple juice (Heo and Uhm, 2017). Some work reported that L. plantarum strains possess organophosphorus pesticide-degrading activity (Li et al., 2018). Therefore, L. plantarum JBE245 was probably an undiscovered PAOs. S. muelleri PUNC, T. turnerae T7901, K. oncopeltii TCC290E, W. endosymbiont TRS, and W. endosymbiont of D. simulans wHa are as intracellular endosymbionts found in specialized cells in organisms (James and Ballard, 2000; Moran et al., 2005; Yang et al., 2009; Alves et al., 2013). They are very likely to ingest phosphate from microbes to maintain the growth. However, these endosymbionts are difficult to appear in the activated sludge. Representatives of the Thiomonas genus are commonly found in moderately acidic (pH 3~5) mine drainage waters (Hallberg and Johnson, 2003; Battaglia-Brunet et al., 2006; Duquesne et al., 2008). Thiomonas strain 3As was isolated from a stream draining an abandoned lead zinc silver mine in the south of France (Duquesne et al., 2008). Like Tm. arsenivorans and Tm. intermediaT (Battaglia-Brunet et al., 2006), strain 3AsT can oxidize As(III) as well as thiosulfate at low pH (Duquesne et al., 2008). In view of the apparently ambiguous available phylogenetic and physiological information regarding strain 3As, whether it has the ability of removing phosphate is not yet known.
The following information can be obtained by analyzing the predicted PAOs. Although Anoxybacillus and Amphibacillus are the members of Bacillus which is considered as PAOs, they are classified separately in MBGD. Through the recognition of A. flavithermus WK1, Anoxybacillus sp. B7M1 b7m1, Anoxybacillus sp. B2M1 b2m1, and A. xylanus NBRC 15112, the accuracy of SVM model is proved indirectly. There are also some microorganisms, just like A. haemolyticum DSM 20595, A. phenanthrenivorans Sphe 3, and C. coralloides DSM 2259, belonging to Actinobacteria or part of gene sequences are the same as Actinobacteria. Some work have been reported that most of the Actinobacteria may be PAOs. Some others are the endosymbionts ingesting P from the hosts to maintain the growth, but not common in activated sludge. It is worth noting that L. plantarum JBE245, belonging to L. plantarum, has the degradation characteristics for organophosphorus. All of them have been predicted by SVM model which can be seen in the Table 1, and the accuracy of the model is proved.
Conclusion
The present work presented a high accuracy SVM algorithm to realize the rapid identification and prediction of PAOs. In this work, all the 6,318 microbial genome sequences were obtained from MBGD. Before the SVM model was established, minimap2 was used to compare the genome sequences. By calculating the similarity between the microbial genome sequences, a matrix of 1*6,318 was achieved. The features were trained by the SVM model, and then they were evaluated with 10-fold cross-validation. The average accuracy we got is 0.9628 ± 0.019. Therefore, the SVM model shows high prediction accuracy and good stability. By predicting 2,652 microorganisms, 22 potential PAOs were obtained. Through the analysis of the predicted potential PAOs, most of them could be indirectly verified their phosphorus removal characteristics from previous reports. This result also further confirmed the accuracy of the model we built. This development illustrated a novel and efficient method to accelerate the screening and identification of microorganism. We also look forward to expanding this work to predict other functional bacteria to provide more possibilities for water treatment.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://mbgd.genome.ad.jp.
Author Contributions
BL wrote the paper and did the experiments. JN provided ideas of this work and supervised this work. XZu revised this manuscript. XZh researched the literature. QX organized the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Nanqi Ren Studio, Academy of Environment & Ecology, Harbin Institute of Technology (Grant No. HSCJ201702) and the National Science and Technology Major Project of Twelfth Five Years (Grant No. 2014ZX07201-012-2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alves, J. M., Serrano, M. G., Maia da Silva, F., Voegtly, L. J., Matveyev, A. V., Teixeira, M. M., et al. (2013). Genome evolution and phylogenomic analysis of candidatus kinetoplastibacterium, the betaproteobacterial endosymbionts of strigomonas and angomonas. Genome Biol. Evol. 5, 338–350. doi: 10.1093/gbe/evt012
Bakolitsa, C., Bateman, A., Jin, K. K., Mcmullan, D., and Axelrod, H. L. (2010). The structure of jann_2411 (duf1470) from Jannaschia sp. at 1.45 Å resolution reveals a new fold (the abate domain) and suggests its possible role as a transcription regulator. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1198–1204. doi: 10.1107/S1744309109025196
Ballabio, C., and Sterlacchini, S. (2012). Support vector machines for landslide susceptibility mapping: the staffora river basin case study, Italy. Math. Geosci. 44, 47–70. doi: 10.1007/s11004-011-9379-9
Banck, G., and Nyman, M. (1986). Tonsillitis and rash associated with Corynebacterium haemolyticum. J. Infect. Dis. 154, 1037–1040. doi: 10.1093/infdis/154.6.1037
Battaglia-Brunet, F., Joulian, C., Garrido, F., Dictor, M. C., Morin, D., and Coupland, K., et al. (2006). Oxidation of arsenite by Thiomonas strains and characterization of Thiomonas arsenivorans sp. nov. Antonie Van Leeuwenhoek. 89, 1–10. doi: 10.1007/s10482-005-9013-2
Beer, M., Kong, Y. H., and Seviour, R. J. (2004). Are some putative glycogen accumulating organisms (GAO) in anaerobic:aerobic activated sludge systems members of the a-Proteobacteria? Microbiology 150, 2267–2275. doi: 10.1099/mic.0.26825-0
Borsodi, A. K., Micsinai, A., Rusznyak, A., Vladar, P., Kovacs, G., Toth, E. M., et al. (2005). Diversity of alkaliphilic and alkalitolerant bacteria cultivated from decomposing reed rhizomes in a Hungarian soda lake. Microb. Ecol. 50, 9–18. doi: 10.1007/s00248-004-0063-1
Cai, W., Huang, W., Lei, Z., Zhang, Z., Lee, D. J., and Adachi, Y. (2019). Granulation of activated sludge using butyrate and valerate as additional carbon source and granular phosphorus removal capacity during wastewater treatment. Bioresour. Technol. 282, 269–274. doi: 10.1016/j.biortech.2019.03.017
Carrier, C., Kalra, A., and Ahmad, S. (2013). Using paleo reconstructions to improve streamflow forecast lead time in the western United States. JAWRA J. Am. Water Resour. Assoc. 49, 1351–1366. doi: 10.1111/jawr.12088
Ch, S., Anand, N., Panigrahi, B. K., and Mathur, S. (2013). Streamflow forecasting by SVM with quantum behaved particle swarm optimization. Neurocomputing. 101, 18–23. doi: 10.1016/j.neucom.2012.07.017
Chaun, H. (2001). Update on the Role of H pylori infection in gastrointestinal disorders. Can. J. Gastroenterol. 15, 251–255. doi: 10.1155/2001/279596
Chen, H., Wang, J., Li, J., and Tang, B. (2017).A texture-based rolling bearing fault diagnosis scheme using adaptive optimal kernel time frequency representation and uniform local binary patterns. Meas. Sci. Technol. 28:035903. doi: 10.1088/1361-6501/aa53a0
Chen, H., Yang, B., Wang, G., Wang, S., Liu, J., and Liu, D. (2012). Support vector machine based diagnostic system for breast cancer using swarm intelligence. J. Med. Syst. 36, 2505–2519. doi: 10.1007/s10916-011-9723-0
Chen, J., Wang, C., and Wang., R. (2008). Combining support vector machines with a pairwise decision tree. IEEE Geosci. Remote Sens. Lett. 5, 409–413. doi: 10.1109/LGRS.2008.916834
Chen, W., Lu, X., Yao, C., Zhu, G., and Xu, Z. (2015). An efficient approach based on bi-sensitivity analysis and genetic algorithm for calibration of activated sludge models. Chem. Eng. J. 259845–259853. doi: 10.1016/j.cej.2014.07.131
Cheng, W., and Jhan, D. (2012). Triaxial accelerometer-based fall detection method using a self-constructing cascade-AdaBoost-SVM classifier. IEEE J. Biomed. Health Inf. 17, 411–419. doi: 10.1109/JBHI.2012.2237034
Clerck, E. D., Vanhoutte, T., Hebb, T., Geerinck, J., and Vos, P. D. (2004). Isolation, Characterization, and Identification of bacterial contaminants in semifinal gelatin extracts. Appl. Environ. Microbiol. 70:3664. doi: 10.1128/AEM.70.6.3664-3672.2004
Collins, M. D., Jones, D., and Schofield, G. M. (1982). Reclassification of 'Corynebacterium haemolyticum' (MacLean, Liebow and Rosenberg) in the genus Arcanobacterium gen.nov. as Arcanobacterium haemolyticum nom.rev. comb.nov. J. Gen. Microbiol. 128, 1279–1281. doi: 10.1099/00221287-128-6-1279
Correll, D. L. (1998). The role of phosphorus in the eutrophication of receiving waters: a review. J. Environ. Qual. 27, 261–266. doi: 10.2134/jeq1998.00472425002700020004x
Deschoenmaeker, F., Facchini, R., Leroy, B., Badri, H., Zhang, C. C., and Wattiez, R. (2014). Proteomic and cellular views of Arthrospira sp. plain 8005 adaptation to nitrogen depletion. Microbiology 160, 1224–1236. doi: 10.1099/mic.0.074641-0
Ding, Y., Tang, J., and Guo, F. (2016). Predicting protein-protein interactions via multivariate mutual information of protein sequences. BMC Bioinformatics 17:398. doi: 10.1186/s12859-016-1253-9
Duquesne, K., Lieutaud, A., Ratouchniak, J., Muller, D., Lett, M. C., and Bonnefoy, V. (2008). Arsenite oxidation by a chemoautotrophic moderately acidophilic Thiomonas sp.: from the strain isolation to the gene study. Environ. Microbiol.10, 228–237. doi: 10.1111/j.1462-2920.2007.01447.x
Fijani, E., Barzegar, R., Deo, R., Tziritis, E., and Skordas, K. (2018). Design and implementation of a hybrid model based on two-layer decomposition method coupled with extreme learning machines to support real-time environmental monitoring of water quality parameters. Sci. Total Environ. 648, 839–853. doi: 10.1016/j.scitotenv.2018.08.221
Fu, W., Tan, J., Zhang, X., Chen, T., and Wang, K. (2019). Blind parameter identification of MAR model and mutation hybrid GWO-SCA optimized SVM for fault diagnosis of rotating machinery. Complexity 2019, 17–34. doi: 10.1155/2019/3264969
Gao, H., Mao, Y., Zhao, X., Liu, W. T., Zhang, T., and Wells, G. (2019). Genome-centric metagenomics resolves microbial diversity and prevalent truncated denitrification pathways in a denitrifying PAO-enriched bioprocess. Water Res. 155, 275–287. doi: 10.1016/j.watres.2019.02.020
Ghaffari, S., Sepahi, A. A., Razavi, M. R., Malekzadeh, F., and Haydarian, H. (2011). Effectiveness of inoculation with isolated Anoxybacillus sp. MGA110 on municipal solid waste composting process. Afr. J. Microbiol. Res. 5, 5373–5378. doi: 10.5897/AJMR11.864
Goh, K. M., Kahar, U. M., Chai, Y. Y., Chong, C. S., Chai, K. P., Ranjani, V., et al. (2013). Recent discoveries and applications of Anoxybacillus. Appl. Microbiol. Biotechnol. 97, 1475–1488. doi: 10.1007/s00253-012-4663-2
Günther, S., Trutnau, M., Kleinsteuber, S., Hause, G., Bley, T., Röske, I., et al. (2009). Dynamics of polyphosphate-accumulating bacteria in wastewater treatment plant microbial communities detected via DAPI (4′,6′-diamidino-2-phenylindole) and tetracycline labeling. Appl. Environ. Microbiol. 75, 2111–2121. doi: 10.1128/AEM.01540-08
Hallberg, K. B., and Johnson, D. B. (2003). Novel acidophiles isolated from moderately acidic mine drainage waters. Hydrometallurgy 71, 139–148. doi: 10.1016/S0304-386X(03)00150-6
He, Z., Wen, X., Liu, H., and Du, J. (2014). A comparative study of artificial neural network, adaptive neuro fuzzy inference system and support vector machine for forecasting river flow in the semiarid mountain region. J. Hydrol. 509, 379–386. doi: 10.1016/j.jhydrol.2013.11.054
Heinen, W., Lauwers, A. M., and Mulders, J. W. M. (1982). Bacillus flavothermus, a newly isolated facultative thermophile. Antonie Van Leeuwenhoek 48, 265–272. doi: 10.1007/BF00400386
Heo, J., and Uhm, T. B. (2017). Complete genome sequence of Lactobacillus Plantarum jbe245 isolated from meju. Korean J. Microbiol. 53, 344–346. doi: 10.7845/kjm.2017.7070
Huang, C., Liu, Q., Li, Z., Ma, X., Hou, Y., and Ren, N., et al. (2020). Relationship between functional bacteria in a denitrification desulfurization system under autotrophic, heterotrophic, and mixotrophic conditions. Water Res. 188:116526. doi: 10.1016/j.watres.2020.116526
Hughes, R. A., and Cornblath, D. R. (2005). Guillain-Barré syndrome. Lancet. 366, 1653–1666. doi: 10.1016/S0140-6736(05)67665-9
Huntley, S., Zhang, Y., Treuner-Lange, A., Kneip, S., Sensen, C. W., and Søgaard-Andersen, L. (2012). Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM 2259. J. Bacteriol. 194, 3012–3013. doi: 10.1128/JB.00397-12
James, A. C., and Ballard, J. W. O. (2000). Expression of cytoplasmic incompatibility in Drosophila simulans and its impact on infection frequencies and distribution of Wolbachia pipientis. Evolution 54, 1661–1672. doi: 10.1111/j.0014-3820.2000.tb00710.x
Janssen, P. J., Morin, N., Mergeay, M., Leroy, B., Wattiez, R., and Vallaeys, T., et al. (2010). Genome sequence of the edible cyanobacterium Arthrospira sp. PCC 8005. J Bacteriol. 192, 2465–2466. doi: 10.1128/JB.00116-10
Ju, X., Hou, J., Tang, Y., Sun, Y., Zheng, S., and Xu, Z. (2016). ZrO2 nanoparticles confined in CMK-3 as highly effective sorbent for phosphate adsorption. Micropor. Mesopor. Mat. 230, 188–195. doi: 10.1016/j.micromeso.2016.05.002
Kallimanis, A., Frillingos, S., Drainas, C., and Koukkou, A. I. (2007). Taxonomic identification, phenanthrene uptake activity and membrane lipid alterations of the PAH degrading Arthrobacter sp. strain Sphe3. Appl. Microbiol. Biotechnol. 76, 709–717. doi: 10.1007/s00253-007-1036-3
Kallimanis, A., Labutti, K. M., Lapidus, A., Clum, A., Lykidis, A., Mavromatis, K., et al. (2011). Complete genome sequence of Arthrobacter phenanthrenivorans type strain (Sphe3). Stand. Genomic Sci. 4, 123–130. doi: 10.4056/sigs.1393494
Kawaharasaki, M., Tanaka, H., Kanagawa, T., and Nakamura, K. (1999). In situ identification of polyphosphate-accumulating bacteria in activated sludge by dual staining with rRNA-targeted oligonucleotide probes and 4′,6-diamidino-2-phenylindol (DAPI) at a polyphosphate-probing concentration. Water Res. 33, 257–265. doi: 10.1016/S0043-1354(98)00183-3
Kong, Y., Nielsen, J. L., and Nielsen, P. H. (2005). Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 71, 4076–4085. doi: 10.1128/AEM.71.7.4076-4085.2005
Kong, Y., Xia, Y., Nielsen, J. L., and Nielsen, P. H. (2007). Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology. 153, 4061–4073. doi: 10.1099/mic.0.2007/007245-0
Li, C., Ma, Y., Mi, Z., Huo, R., Zhou, T., and Hai, H., et al. (2018). Screening for Lactobacillus plantarum strains that possess organophosphorus pesticide-degrading activity and metabolomic analysis of phorate degradation. Front. Microbiol. 9:2048. doi: 10.3389/fmicb.2018.02048
Li, H. (2016). Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110. doi: 10.1093/bioinformatics/btw152
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191
Lin, Z., Wang, Y., Huang, W., Wang, J., Chen, L., Zhou, J., et al. (2019). Single-stage denitrifying phosphorus removal biofilter utilizing intracellular carbon source for advanced nutrient removal and phosphorus recovery. Bioresour. Technol. 277, 27–36. doi: 10.1016/j.biortech.2019.01.025
Linder, R. (1997). Rhodococcus equi and Arcanobacterium haemolyticum: two “coryneform” bacteria increasingly recognized as agents of human infection. Emerging Infect. Dis. 3, 145–153. doi: 10.3201/eid0302.970207
Litke, D. W. (1999). Review of Phosphorus Control Measures in the United States and Their Effects on Water Quality. Denver, CO: U.S. Geological Survey.
Liu, B., Nan, J., Zu, X., Zhang, X., Huang, W., and Wang, W. (2020). La-based-adsorbents for efficient biological phosphorus treatment of wastewater: synergistically strengthen of chemical and biological removal. Chemosphere 255:127010. doi: 10.1016/j.chemosphere.2020.127010
Liu, M., and Lu, J. (2014). Support vector machine-an alternative to artificial neuron network for water quality forecasting in an agricultural nonpoint source polluted river? Environ. Sci. Pollut. R. 21, 11036–11053. doi: 10.1007/s11356-014-3046-x
Loganathan, P., Vigneswaran, S., Kandasamy, J., and Bolan, N. S. (2014). Removal and recovery of phosphate from water using sorption. Crit. Rev. Environ. Sci. Technol. 44, 847–907. doi: 10.1080/10643389.2012.741311
Mackenzie, A., Fuite, L. A., Chan, F. T., King, J., Allen, U., MacDonald, N., et al. (1995). Incidence and pathogenicity of Arcanobacterium haemolyticum during a 2-year study in Ottawa. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 21, 177–181. doi: 10.1093/clinids/21.1.177
Miller, R. A., Brancato, F., and Holmes, K. K. (1986). Corynebacterium hemolyticum as a cause of pharyngitis and scarlatiniform rash in young adults. Ann. Intern. Med. 105, 867–872. doi: 10.7326/0003-4819-105-6-867
Moran, N., Tran, P., and Gerardo, N. (2005). Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum bacteroidetes. Appl. Environ. Biol. 27. 8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005
Mountain, B. W., Benning, L. G., and Boerema, J. A. (2003). Experimental studies on New Zealand hot spring sinters: rates of growth and textural development. Can. J. Earth Sci. 40, 1643–1667. doi: 10.1139/e03-068
Nguyen, H. T., Le, V. Q., Hansen, A. A., Nielsen, J. L., and Nielsen, P. H. (2011). High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems. FEMSMicrobiol. Ecol. 76, 256–267. doi: 10.1111/j.1574-6941.2011.01049.x
Nguyen, H. T., Nielsen, J. L., and Nielsen, P. H. (2012). “Candidatus halomonas phosphatis”, a novel polyphosphate-accumulating organism in full-scale enhanced biological phosphorus removal plants. Environ. Microbiol. 14, 2826–2837. doi: 10.1111/j.1462-2920.2012.02826.x
Niimura, Y., Koh, E., Yanagida, F., Suzuki, K.-I., Komagata, K., and, Kozaki, M. (1990). Amphibacillus xylanus gen. nov., sp. nov., a facultatively anaerobic sporeforming xylan-digesting bacterium which lacks cytochrome, quinone, and catalase. Int. J. Syst. Bacteriol. 40, 297–301. doi: 10.1099/00207713-40-3-297
Pan, G., Jiang, L., Tang, J., and Guo, F. (2018). A novel computational method for detecting DNA methylation sites with DNA sequence information and hysicochemical properties. Int. J. Mol. Sci. 19:511. doi: 10.3390/ijms19020511
Pang, J., Yang, S., He, L., Chen, Y., Cao, G., Zhao, L., et al. (2019). An influent responsive control strategy withmachine learning: Q-learning based optimization method for a biological phosphorus removal system. Chemosphere 234, 893–901. doi: 10.1016/j.chemosphere.2019.06.103
Parker, C. T., Huynh, S., Heikema, A. P., Cooper, K. K., and Miller, W. G. (2015). Complete genome sequences of Campylobacter jejuni strains RM3196 (233.94) and RM3197 (308.95) isolated from patients with guillain-barré syndrome. Genome Announc. 3, e01312–e01315. doi: 10.1128/genomeA.01312-15
Pawlowski, A. C., Wang, W., Koteva, K., Barton, H. A., Mcarthur, A. G., and Wright, G. D. (2016). A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun. 7:13803. doi: 10.1038/ncomms13803
Peng, J., Yang, J., and Jin, Q. (2009). The molecular evolutionary history of Shigella spp. and enteroinvasive Escherichia coli. Infect. Genetics Evolut. 9, 147–152. doi: 10.1016/j.meegid.2008.10.003
Peng, J., and Zhao, T. (2020). Reduction in TOM1 expression exacerbates Alzheimer's disease. Proc. Natl. Acad. Sci. 117, 3915–3916. doi: 10.1073/pnas.1917589117
Pham, B. T., Jaafari, A., Prakash, I., and Bui, D. T. (2018). A novel hybrid intelligent model of support vector machines and the multiboost ensemble for landslide susceptibility modeling. Bull. Eng. Geol. Environ. 78, 2865–2886. doi: 10.1007/s10064-018-1281-y
Pikuta, E., Lysenko, A., Chuvilskaya, N., Mendrock, U., Hippe, H., and Suzina, N., et al. (2000). Anoxybacillus pushchinensis gen. nov. sp. nov. a novel anaerobic, alkaliphilic, moderately thermophilic bacterium from manure, and description of Anoxybacillus flavitherms comb. nov. Int. J. Syst. Evolut. Microbiol. 50, 2109–2117. doi: 10.1099/00207713-50-6-2109
Roberts, M., Hayes, W., Hunt, B., Mount, S., and Yorke, J. (2004). Reducing storage requirements for biological sequence comparison. Bioinformatics.20, 3363–3369. doi: 10.1093/bioinformatics/bth408
Rueckert, A., Ronimus, R. S., and Morgan, H. W. (2005). Development of a rapid detection and enumeration method for thermophilic bacilli in milk powders. J. Microbiol. Methods 60, 155–167. doi: 10.1016/j.mimet.2004.09.008
Salehi, S., Cheng, K. Y., Heitz, A., and Ginige, M. P. (2019). A novel storage driven granular post denitrification process: long-term effects of volume reduction on phosphate recovery. Chem. Eng. J. 356, 534–542. doi: 10.1016/j.cej.2018.08.139
Senior, K. (2009). Estimating the global burden of foodborne disease. Lancet Infect. Dis. 9, 80–81. doi: 10.1016/S1473-3099(09)70008-8
Shi, H., and Lee, C. (2006). Combining anoxic denitrifying ability with aerobicanoxic phosphorus-removal examinations to screen denitrifying phosphorus-removing bacteria. Int. Biodeterior. Biodegradation. 57, 121–128. doi: 10.1016/j.ibiod.2006.01.001
Shi, P., Liang, K., Han, D., and Zhang, Y. (2017). A novel intelligent fault diagnosis method of rotating machinery based on deep learning and PSO-SVM. J. Vibroeng. 19, 5932–5946. doi: 10.21595/jve.2017.18380
Shi, Y., Chai, L., Yang, Z., Jing, Q., Chen, R., and Chen, Y. (2012). Identification and hexavalent chromium reduction characteristics of Pannonibacter phragmitetus. Bioprocess Biosyst. Eng. 35, 843–850. doi: 10.1007/s00449-011-0668-y
Smith, V. H., Tilman, G. D., and Nekola, J. C. (1999). Eutrophication: impacts of excess nutrient inputs on freshwater, marine, and terrestrial ecosystems. Environ. Pollut. 100, 179–196. doi: 10.1016/S0269-7491(99)00091-3
Song, Z., Liu, Q., Guo, H., Ju, R., Zhao, Y., and Li, J., et al. (2012). Tostadin, a novel antibacterial peptide from an antagonistic microorganism Brevibacillus brevis xdh. Bioresour. Technol. 111, 504–506. doi: 10.1016/j.biortech.2012.02.051
Stoddard, J. L., Van Sickle, J., Herlihy, A. T., Brahney, J., Paulsen, S., and Peck, D. V., et al. (2016). Continental-scale increase in lake and stream phosphorus: are oligotrophic systems disappearing in the United States? Environ. Sci. Technol. 50, 3409–3415. doi: 10.1021/acs.est.5b05950
Sun, L., Cao, W., Zhang, H., Dong, Y., Zhang, J., Tu, B., et al. (2016). Optimal growth conditions and nutrient removal characteristic of a denitrifying phosphorus-accumulating organism. Desalin Water Treat. 57, 25028–25035. doi: 10.1080/19443994.2016.1144531
Tsuneda, S., Miyauchi, R., Ohno, T., and Hirata, A. (2005). Characterization of denitrifying polyphosphate-accumulating organisms in activated sludge based on nitrite reductase gene. J. Biosci. Bioeng. 99, 403–407. doi: 10.1263/jbb.99.403
Uchiyama, I., Mihara, M., Nishide, H., and Chiba, H. (2013). MBGD update 2013: the microbial genome database for exploring the diversity of microbial world. Nucleic Acids Res. 41, D631–D635. doi: 10.1093/nar/gks1006
Vapnik, V. N. (1995). The Nature of Statistical Learning Theory. New York, NY: Springer Verlag. doi: 10.1007/978-1-4757-2440-0
Wang, D., Li, X., Yang, Q., Zheng, W., Cao, J., Zeng, G., et al. (2009). Effect and mechanism of carbon sources on phosphorus uptake by microorganisms in sequencing batch reactors with the single-stage oxic process. Sci. China Series B Chem. 52, 2358–2365. doi: 10.1007/s11426-009-0152-6
Wang, M., Zhang, X., Jiang, T., Hu, S., Yi, Z., Zhou, Y., et al. (2017). Liver abscess caused by Pannonibacter phragmitetus: case report and literature review. Front. Med. 4:48. doi: 10.3389/fmed.2017.00048
Wang, X., Zhao, J., Yu, D., Du, S., Yuan, M., and Zhen, J. (2019). Evaluating the potential for sustaining mainstream anammox by endogenous partial denitrification and phosphorus removal for energy-efficient wastewater treatment. Bioresour. Technol. 284, 302–314. doi: 10.1016/j.biortech.2019.03.127
Wang, Y., Yang, Z., Peng, B., Chai, L., Wu, B., and Wu, R. (2013). Biotreatment of chromite ore processing residue by Pannonibacter phragmitetus BB. Environ. Sci. Pollut. Res. 20, 5593–5602. doi: 10.1007/s11356-013-1526-z
Xu, L., Luo, M., Yang, L., Wei, X., Lin, X., and Liu, H. (2011). Encapsulation of Pannonibacter phragmitetus LSSE-09 in alginate-carboxymethyl cellulose capsules for reduction of hexavalent chromium under alkaline conditions. J. Ind. Microbiol. Biotechnol. 38, 1709–1718. doi: 10.1007/s10295-011-0960-5
Xu, X., Qiu, L., Wang, C., and Yang, F. (2019). Achieving mainstream nitrogen and phosphorus removal through simultaneous partial nitrification, anammox, denitrification, and denitrifying phosphorus removal (SNADPR) process in a single-tank integrative reactor. Bioresour. Technol. 284, 80–89. doi: 10.1016/j.biortech,.2019.03.109
Yang, J. C., Madupu, R., Durkin, a, S., Ekborg, N a, Pedamallu, C. S., Hostetler, J. B., et al. (2009). The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (Shipworms). PLoS ONE 4:e6085. doi: 10.1371/journal.pone.0006085
Zeng, L., Li, X., and Liu, J. (2004). Adsorptive removal of phosphate from aqueous solutions using iron oxide tailings. Water Res. 38, 1318–1326. doi: 10.1016/j.watres.2003.12.009
Zhao, T., Hu, Y., and Cheng, L. (2020a). Deep-drm: a computational method for identifying disease-related metabolites based on graph deep learning approaches. Brief. Bioinformatics. bbaa212. doi: 10.1093/bib/bbaa212
Zhao, T., Hu, Y., Valsdottir, L. R., Zang, T., and Peng, J. (2020b). Identifying drug–target interactions based on graph convolutional network and deep neural network. Brief. Bioinformatics. bbaa044. doi: 10.1093/bib/bbaa044
Zhao, T., Hu, Y., Zang, T., and Cheng, L. (2020c). Mrtfb regulates the expression of nomo1 in colon. Proc. Natl. Acad. Sci. U.S.A. 117:202000499. doi: 10.1073/pnas.2000499117
Zhao, T., Lyu, S., Lu, G., Juan, L., Zeng, X., and Wei, Z., et al. (2020d). SC2disease: a manually curated database of single-cell transcriptome for human diseases. Nucleic Acids Research. gkaa838. doi: 10.1093/nar/gkaa838
Zhu, Y., Chen, K., Ding, Y., Situ, D., Li, Y., and Long, Y., et al. (2019). Metabolic and proteomic mechanism of benzo[a]pyrene degradation by brevibacillus brevis. Ecotoxicol. Environ. Saf. 172, 1–10. doi: 10.1016/j.ecoenv.2019.01.044
Keywords: polyphosphate-accumulating organisms, genome sequences, minimap2, support vector machine, prediction
Citation: Liu B, Nan J, Zu X, Zhang X and Xiao Q (2021) Identification of Genome Sequences of Polyphosphate-Accumulating Organisms by Machine Learning. Front. Cell Dev. Biol. 8:626221. doi: 10.3389/fcell.2020.626221
Received: 05 November 2020; Accepted: 15 December 2020;
Published: 18 January 2021.
Edited by:
Lei Deng, Central South University, ChinaReviewed by:
Sheng Li, Wuhan University, ChinaYang Yang, Inner Mongolia University, China
Fei Shen, Guangzhou First People's Hospital, China
Copyright © 2021 Liu, Nan, Zu, Zhang and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Nan, nanjun119@163.com
 Bohan Liu
Bohan Liu  Jun Nan
Jun Nan Xinhui Zhang
Xinhui Zhang