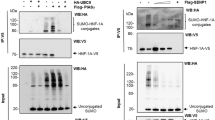
Abstract
Mutations in hepatocyte nuclear factor (HNF)1A gene cause the most common form of Maturity-onset diabetes of the young (MODY), a monogenic subtype of diabetes mellitus. Functional characterization of mutant proteins reveals that mutations may disrupt DNA binding capacity, transactivation ability and nuclear localization of HNF1A depending on the position of the mutation. Previously identified Arg271Trp and Ser345Tyr mutations in HNF1A were found to be defective in nuclear localization. Arg271 residue resides in a region similar to classical nuclear localization signal (NLS) motif, while Ser345 does not. Importin α family members recognize NLS motifs on cargo proteins and subsequently translocate them into nucleus. Here, we first investigated the nuclear localization mechanism of wild type HNF1A protein. For this purpose, we analyzed the interaction of HNF1A with three mouse homolog importin α proteins (KPNA2, KPNA4 and KPNA6) by co-immunoprecipitation assay and molecular docking simulation. Hereby, KPNA6 was identified as the main import receptor, which is responsible for the transport of HNF1A into the nucleus. Immunolocalization studies in mouse pancreatic cells (Min6) also confirmed the co-localization of HNF1A and KPNA6 in the cytoplasm. Secondly, the interaction between KPNA6 and mutant HNF1A proteins (Arg271Trp and Ser345Tyr) was assessed. Co-immunoprecipitation studies revealed a reduced interaction compared to wild type HNF1A. Our study demonstrated for the first time that HNF1A transcription factor is recognized and transported by importin/karyopherin import family, and mutations in NLS motifs may disrupt the interaction leading to nuclear localization abnormalities and MODY phenotype.






Similar content being viewed by others
References
Murphy R, Ellard S, Hattersley AT (2008) Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab. https://doi.org/10.1038/ncpendmet0778
Fajans SS, Bell GI (2011) MODY: history, genetics, pathophysiology, and clinical decision making. Diabetes Care. https://doi.org/10.2337/dc11-0035
Anık A, Çatlı G, Abacı A, Böber E (2014) Maturity-onset diabetes of the young (MODY): an update. J Pediatr Endocrinol Metab. https://doi.org/10.1515/jpem-2014-0384
Urakami T (2019) Maturity-onset diabetes of the young (MODY): current perspectives on diagnosis and treatment. Diabetes Metab Syndr Obes. https://doi.org/10.2147/DMSO.S179793
Wang H, Antinozzi PA, Hagenfeldt KA, Maechler P, Wollheim CB (2000) Molecular targets of a human HNF1 alpha mutation responsible for pancreatic beta-cell dysfunction. EMBO J. https://doi.org/10.1093/emboj/19.16.4257
Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science. https://doi.org/10.1126/science.1089769
Servitja JM, Pignatelli M, Maestro MA, Cardalda C, Boj SF, Lozano J, Blanco E, Lafuente A, McCarthy MI, Sumoy L, Guigó R, Ferrer J (2009) Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol Cell Biol. https://doi.org/10.1128/MCB.01389-08
Pontoglio M, Sreenan S, Roe M, Pugh W, Ostrega D, Doyen A, Pick AJ, Baldwin A, Velho G, Froguel P, Levisetti M, Bonner-Weir S, Bell GI, Yaniv M, Polonsky KS (1998) Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J Clin Invest. https://doi.org/10.1172/JCI2548
Mendel DB, Crabtree GR (1991) HNF-1, a member of a novel class of dimerizing homeodomain proteins. J Biol Chem 266(2):677–680
Valkovicova T, Skopkova M, Stanik J, Gasperikova D (2019) Novel insights into genetics and clinics of the HNF1A-MODY. Endocr Regul. https://doi.org/10.2478/enr-2019-0013
Bjørkhaug L, Sagen JV, Thorsby P, Søvik O, Molven A, Njølstad PR (2003) Hepatocyte nuclear factor-1 alpha gene mutations and diabetes in Norway. J Clin Endocrinol Metab. https://doi.org/10.1210/jc.2002-020945
Najmi LA, Aukrust I, Flannick J et al (2017) Functional investigations of HNF1A identify rare variants as risk factors for type 2 diabetes in the general population. Diabetes. https://doi.org/10.2337/db16-0460
Bjørkhaug L, Bratland A, Njølstad PR, Molven A (2005) Functional dissection of the HNF-1alpha transcription factor: a study on nuclear localization and transcriptional activation. DNA Cell Biol. https://doi.org/10.1089/dna.2005.24.661
Radu A, Blobel G, Moore MS (1995) Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.92.5.1769
Li C, Goryaynov A, Yang W (2016) The selective permeability barrier in the nuclear pore complex. Nucleus. https://doi.org/10.1080/19491034.2016.1238997
Mosammaparast N, Pemberton LF (2004) Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. https://doi.org/10.1016/j.tcb.2004.09.004
Freitas N, Cunha C (2009) Mechanisms and signals for the nuclear import of proteins. Curr Genomics. https://doi.org/10.2174/138920209789503941
Ström AC, Weis K (2001) Importin-beta-like nuclear transport receptors. Genome Biol. https://doi.org/10.1186/gb-2001-2-6-reviews3008
Lange A, McLane LM, Mills RE, Devine SE, Corbett AH (2010) Expanding the definition of the classical bipartite nuclear localization signal. Traffic. https://doi.org/10.1111/j.1600-0854.2009.01028.xn
Cros JF, García-Sastre A, Palese P (2005) An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic. https://doi.org/10.1111/j.1600-0854.2005.00263.x
Pumroy RA, Cingolani G (2015) Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem J. https://doi.org/10.1042/BJ20141186
Galán M, García-Herrero CM, Azriel S, Gargallo M, Durán M, Gorgojo JJ, Andía VM, Navas MA (2011) Differential effects of HNF-1α mutations associated with familial young-onset diabetes on target gene regulation. Mol Med. https://doi.org/10.2119/molmed.2010.00097
Chi YI, Frantz JD, Oh BC, Hansen L, Dhe-Paganon S, Shoelson SE (2002) Diabetes mutations delineate an atypical POU domain in HNF-1alpha. Mol Cell. https://doi.org/10.1016/s1097-2765(02)00704-9
Hirano H, Matsuura Y (2011) Sensing actin dynamics: structural basis for G-actin-sensitive nuclear import of MAL. Biochem Biophys Res Commun. https://doi.org/10.1016/j.bbrc.2011.09.079
Bienert S, Waterhouse A, de Beer TAP, Tauriello G, Studer G, Bordoli L, Schwede T (2017) The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw1132
Koyama M, Matsuura Y (2017) Crystal structure of importin-α3 bound to the nuclear localization signal of Ran-binding protein 3. Biochem Biophys Res Commun. https://doi.org/10.1016/j.bbrc.2017.07.155
Pumroy RA, Ke S, Hart DJ, Zachariae U, Cingolani G (2015) Molecular determinants for nuclear import of influenza A PB2 by importin α isoforms 3 and 7. Structure. https://doi.org/10.1016/j.str.2014.11.015
Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z (2014) ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics. https://doi.org/10.1093/bioinformatics/btu097
Mintseris J, Pierce B, Wiehe K, Anderson R, Chen R, Weng Z (2007) Integrating statistical pair potentials into protein complex prediction. Proteins. https://doi.org/10.1002/prot.21502
Krieger E, Koraimann G, Vriend G (2002) Increasing the precision of comparative models with YASARA NOVA–a self-parameterizing force field. Proteins. https://doi.org/10.1002/prot.10104
Oka M, Yoneda Y (2018) Importin α: functions as a nuclear transport factor and beyond. Proc Jpn Acad Ser B. https://doi.org/10.2183/pjab.94.018
Melen K, Fagerlund R, Franke J, Kohler M, Kinnunen L, Julkunen I (2003) Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem. https://doi.org/10.1074/jbc.M303571200
Köhler M, Buchwalow IB, Alexander G, Christiansen M, Shagdarsuren E, Samoilova V, Hartmann E, Mervaala EM, Haller H (2001) Increased importin alpha protein expression in diabetic nephropathy. Kidney Int. https://doi.org/10.1046/j.1523-1755.2001.00069.x
Balamurugan K, Bjørkhaug L, Mahajan S, Kanthimathi S, Njølstad PR, Srinivasan N, Mohan V, Radha V (2016) Structure-function studies of HNF1A (MODY3) gene mutations in South Indian patients with monogenic diabetes. Clin Genet. https://doi.org/10.1111/cge.12757
Nakada R, Hirano H, Matsuura Y (2015) Structure of importin-α bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci Rep. https://doi.org/10.1038/srep15055
Acknowledgements
We acknowledge the funding support from the Scientific and Technological Research Council of Turkey (TÜBİTAK) (Grant Number: 116Z564)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fareed, F.M.A., Korulu, S., Özbil, M. et al. HNF1A-MODY Mutations in Nuclear Localization Signal Impair HNF1A-Import Receptor KPNA6 Interactions. Protein J 40, 512–521 (2021). https://doi.org/10.1007/s10930-020-09959-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09959-0