Abstract
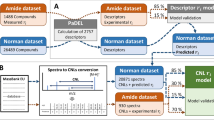
To investigate the structure-dependent peptide mobility behavior in ion mobility spectrometry (IMS), quantitative structure-spectrum relationship (QSSR) is systematically modeled and predicted for the collision cross section Ω values of totally 162 single-protonated tripeptide fragments extracted from the Bacillus subtilis lipase A. Two different types of structure characterization methods, namely, local and global descriptor as well as three machine learning methods, namely, partial least squares (PLS), support vector machine (SVM) and Gaussian process (GP), are employed to parameterize and correlate the structures and Ω values of these peptide samples. In this procedure, the local descriptor is derived from the principal component analysis (PCA) of 516 physicochemical properties for 20 standard amino acids, which can be used to sequentially characterize the three amino acid residues composing a tripeptide. The global descriptor is calculated using CODESSA method, which can generate > 200 statistically significant variables to characterize the whole molecular structure of a tripeptide. The obtained QSSR models are evaluated rigorously via tenfold cross-validation and Monte Carlo cross-validation (MCCV). A comprehensive comparison is performed on the resulting statistics arising from the systematic combination of different descriptor types and machine learning methods. It is revealed that the local descriptor-based QSSR models have a better fitting ability and predictive power, but worse interpretability, than those based on the global descriptor. In addition, since the QSSR modeling using local descriptor does not consider the three-dimensional conformation of tripeptide samples, the method would be largely efficient as compared to the global descriptor.


Similar content being viewed by others
References
Verbeck GF, Ruotolo BT, Sawyer HA, Gillig KJ, Russell DH (2002) A fundamental introduction to ion mobility mass spectrometry applied to the analysis of biomolecules. J Biomol Tech 13:56–61
Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE (2008) Biomolecule analysis by ion mobility spectrometry. Annu Rev Anal Chem. 1:101–1035
Collins DC, Lee ML, Fresenius J (2001) Electrospray ionization gas-phase electrophoresis under ambient conditions and its potential or high-speed separations. Anal Chem 369:225–233
McLean JA, Russell DH (2003) Sub-femtomole peptide detection in ion mobility-time-of-flight mass spectrometry measurements. J Proteome Res 2:427–430
Myung S, Lee YJ, Moon MH, Taraszka J, Sowell R, Koeniger S, Hilderbrand AE, Valentine SJ, Cherbas L, Cherbas P, Kaufmann TC, Miller DF, Mechref Y, Novotny MV, Ewing MA, Sporleder CR, Clemmer DE (2003) Development of high-sensitivity ion trap ion mobility spectrometry time-of-flight techniques: a high-throughput nano-LC-IMS-TOF separation of peptides arising from a Drosophila protein extract. Anal Chem 75:5137–5145
Dwivedi P, Wu P, Klopsch SJ, Puzon GJ, Xun L, Hill HH (2008) Metabolic profiling by ion mobility mass spectrometry (IMMS). Metabolomics 4:63–80
Shvartsburg AA, Jarrold MF (1996) An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem Phys Lett 261:86–91
Revercomb HE, Mason EA (1975) Theory of plasma chromatography/gaseous electrophoresis: a review. Anal Chem 47:970–983
Counterman AE, Clemmer DE (1999) Volumes of individual amino acid residues in gas-phase peptide ions. J Am Chem Soc 121:4031–4039
Valentine SJ, Counterman AE, Hoaglund-Hyzer CS, Clemmer DE (1999) Intrinsic amino acid size parameters from a series of 113 lysine-terminated tryptic digest peptide ions. J Phys Chem B 103:1203–1207
Shvartsburg AA, Siu KWM (2001) Prediction of peptide ion mobilities via a priori calculations from intrinsic size parameters of amino acid residues. J Am Soc Mass Spectrom 12:885–888
Mosier PD, Counterman AE, Jurs PC (2002) Prediction of peptide ion collision cross sections from topological molecular structure and amino acid parameters. Anal Chem 74:1360–1370
Zhou P, Zeng H, Tian FF, Li B, Li ZL (2007) Applying novel molecular electronegativity-interaction vector (MEIV) to QSPR study on collision cross section of singly protonated peptides. QSAR Comb Sci 26:117–121
Zhou P, Tian FF, Li ZL (2007) Quantitative structure–property relationship studies for collision cross sections of 579 singly protonated peptides based on a novel descriptor as molecular graph fingerprint (MoGF). Anal Chim Acta 597:214–222
Zhou P, Tian F, Wu Y, Li Z, Shang Z (2008) Quantitative sequence–activity model (QSAM): applying QSAR strategy to model and predict bioactivity and function of peptides, proteins and nucleic Acids. Curr Comput Aided Drug Des 4:311–321
Hilderbrand AE, Clemmer DE (2005) Determination of sequence-specific intrinsic size parameters from cross sections for 162 tripeptides. J Phys Chem B 109:11802–11809
Nakai K, Kidera A, Kanehisa M (1988) Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng 2:93–100
Kawashima S, Ogata H, Kanehisa M (1999) AAindex: amino acid index database. Nucleic Acids Res 27:368–369
Lu Y, Bulka B, DesJardins M, Freeland SJ (2007) Amino acid quantitative structure property relationship database: a webbased platform for quantitative investigations of amino acids. Protein Eng Des Sel 20:347–351
Kawashima S, Pokarowski P, Pokarowska M, Kolinski A, Katayama T, Kanehisa M (2008) AAindex: amino acid index database, progress report. Nucleic Acids Res 36:D202–D205
Du H, Zhang X, Wang J, Yao X, Hu Z (2008) Novel approaches to predict the retention of histidine-containing peptides in immobilized metal-affinity chromatography. Proteomics 8:2185–2195
Katritzky AR, Lobanov VS, Karelson M (1995) QSPR: the correlation and quantitative prediction of chemical and physical properties from structure. Chem Soc Rev 24:279–287
Wold S, Sjöström M, Eriksson L (2001) PLS regression: a basic tool of chemometrics. Chemometr Intell Lab Syst 58:109–130
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Zhou P, Chen X, Wu Y, Shang Z (2010) Gaussian process: an alternative approach for QSAM modeling of peptides. Amino Acids 38:199–212
Zhou Y, Ni Z, Chen KP, Liu HJ, Chen L, Lian CQ, Yan VLR (2013) Modeling protein–peptide recognition based on classical quantitative structure–affinity relationship approach: implication for proteome-wide inference of peptide-mediated interactions. Protein J 32(7):568–578
Obrezanova O, Csanyi G, Gola JMR, Segall MD (2007) Gaussian processes: a method for automatic QSAR modeling of ADME properties. J Chem Inf Model 47:1847–1857
Wolfe P (1969) Convergence conditions for ascent methods. SIAM Rev 11:226–235
Xu QS, Liang YZ (2001) Monte Carlo cross validation. Chemometr Intell Lab Syst 56:1–11
Manchester J, Czerminski R (2008) SAMFA: simplifying molecular description for 3D-QSAR. J Chem Inf Model 48:167–1173
Zhou P, Tian F, Lv F, Shang Z (2009) Comprehensive comparison of eight statistical modelling methods used in quantitative structure–retention relationship studies for liquid chromatographic retention times of peptides generated by protease digestion of the Escherichia coli proteome. J Chromatogr A 1216:3107–3116
Yan J, Cao DS, Guo FQ et al (2012) Comparison of quantitative structure-retention relationship models on four stationary phases with different polarity for a diverse set of flavor compounds. J Chromatogr A 1223:118–125
Acknowledgements
This work was supported by the Jiangsu University for Advanced Professionals (No. 1281330021), and Open Funding Project of the State Key Laboratory of Biochemical Engineering, China. (2018KF-02)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ni, Z., Wang, A., Kang, L. et al. QSSR Modeling of Bacillus Subtilis Lipase A Peptide Collision Cross-Sections in Ion Mobility Spectrometry: Local Descriptor Versus Global Descriptor. Protein J 40, 54–62 (2021). https://doi.org/10.1007/s10930-020-09960-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09960-7