Abstract
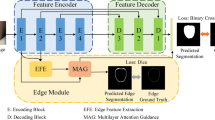
In order to solve the problems of low gray scale contrast and blurred organ boundaries in some medical images, we proposed a joint algorithm of Multi-Attention Parallel CNNs and Independent Recurrent Neural Networks (MACIR) with word embedding technique combined. First, the word embedding technique is used to map the sparse spatial relation matrix into a real dense vector, which is combined with gray scale and edge matrix as input features. The multi -attention mechanism is used to add weight information to capture the importance of each feature more sensitive. Then, the Parallel Convolutional Neural Networks are used to fully exploit the deep semantic information, and IndRNN is introduced to avoid the loss of pixel hierarchy information and realize the integration of information flow. Finally, the Softmax classifier is used to complete the medical image segmentation task. Experiments showed that word embedding MACIR algorithm could effectively improve the segmentation performance of medical images on the data sets of lung X-ray and cervical CT images.





Similar content being viewed by others
REFERENCES
Hwang, S. and Park, S., Accurate lung segmentation via network-wise training of convolutional networks, in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Cham: Springer, 2017.
Tian Juanxiu, Liu Guocai, Gu Shanshan, et al., Research and challenge of medical image analysis deep learning method, J. Autom., 2018, vol. 44, no. 3, pp. 401–424.
LeCun, Y., Bengio, Y., and Hinton, G., Deep learning, Nature, 2015, vol. 521, no. 7553, p. 436.
Ma Chao, Liu Yashu, Luo Gongning, et al., 3D MR image segmentation based on cascaded random forest and active contours, J. Autom., 2019, vol. 45, no. 5, pp. 1004–1014.
Li Xiangxia, Li Bin, Tian Lianfang, et al., Segmentation of ground glass-type pulmonary nodules based on sparse representation and random walk, J. Autom., 2018, vol. 44, no. 9, pp. 1637–1647.
Onoma, D.P., Ruan, S., Thureau, S., et al., Segmentation of heterogeneous or small FDG PET positive tissue based on a 3D-locally adaptive random walk algorithm, Comput. Med. Imaging Graphics, 2014, vol. 38, no. 8, pp. 753–763.
Garnavi, R., Aldeen, M., Celebi, M.E., et al., Border detection in dermoscopy images using hybrid thresholding on optimized color channels, Comput. Med. Imaging Graphics, 2011, vol. 35, no. 2, pp. 105–115.
Ge, Z., Demyanov, S., Bozorgtabar, B., et al., Exploiting local and generic features for accurate skin lesions classification using clinical and dermoscopy imaging, 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), 2017, pp. 986–990.
Abuzaghleh, O., Barkana, B.D., and Faezipour, M., Automated skin lesion analysis based on color and shape geometry feature set for melanoma early detection and prevention, IEEE Long Island Systems, Applications and Technology (LISAT) Conference 2014, 2014, pp. 1–6.
Akbulut, Y., Guo, Y., Sengür, A., et al., An effective color texture image segmentation algorithm based on hermite transform, Appl. Soft Comput., 2018, vol. 67, pp. 494–504.
Tahir, B., Iqbal, S., Usman, Ghani., Khan, M., et al., Feature enhancement framework for brain tumor segmentation and classification, Microsc. Res. Tech., 2019, vol. 82, no. 6, pp. 803–811.
Barui, S., Latha, S., Samiappan, D., et al., SVM pixel classification on colour image segmentation, J. Phys.: Conf. Ser., 2018, vol. 1000.
Chan, Y.H., Zeng, Y.Z., Wu, H.C., et al., Effective pneumothorax detection for chest X-ray images using local binary pattern and support vector machine, J. Healthcare Eng., 2018, vol. 2018.
Long, J., Shelhamer, E., and Darrell, T., Fully convolutional networks for semantic segmentation, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, pp. 3431–3440.
Ronneberger, O., Fischer, P., and Brox, T., U-net: Convolutional networks for biomedical image segmentation, International Conference on Medical Image Computing and Computer-Assisted Intervention, Cham: Springer, 2015, pp. 234–241.
Li, Z., Gan, Y., Liang, X., et al., LSTM-CF: Unifying context modeling and fusion with LSTMS for RGB-d scene labeling, European Conference on Computer Vision, Cham: Springer, 2016, pp. 541–557.
Wang, X., Girshick, R., Gupta, A., et al., Non-local neural networks, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 7794–7803.
Wang, S., Zhou, M., Liu, Z., et al., Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation, Med. Image Anal., 2017, vol. 40, pp. 172–183.
Goldberg, Y. and Levy, O., Word2vec explained: Deriving Mikolov et al.'s negative-sampling word-embedding method, 2014. arXiv:1402.3722.
Henry, S., Cuffy, C., and McInnes, B.T., Vector representations of multi-word terms for semantic relatedness, J. Biomed. Inf., 2018, p. 77.
Bamler, R. and Mandt, S., Dynamic word embeddings, Proceedings of the 34th International Conference on Machine Learning, 2017, vol. 70, pp. 380–389.
Mikolov, T., Karafiát, M., Burget, L., et al., Recurrent neural network based language model, Eleventh Annual Conference of the International Speech Communication Association, 2010.
Li, S., Li, W., Cook, C., et al., Independently recurrent neural network (INDRNN): Building a longer and deeper RNN, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 5457–5466.
Van Ginneken, B., Stegmann, M.B., and Loog, M., Segmentation of anatomical structures in chest radiographs using supervised methods: A comparative study on a public database, Med. Image Anal., 2006, vol. 10, no. 1, pp. 19–40.
Clark, K., Vendt, B., Smith, K., et al., The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository, J. Digital Imaging, 2013, vol. 26, no. 6, pp. 1045–1057.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (no. 61765014); Reserve Talents Project of National High-level Personnel of Special Support Program (QN2016YX0324); Urumqi Science and Technology Project (nos. P161310002 and Y161010025); and Reserve Talents Project of National High-level Personnel of Special Support Program (Xinjiang [2014]22).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflicts of interest.
About this article
Cite this article
Junlong Cheng, Tian, S., Yu, L. et al. Multi-Attention Mechanism Medical Image Segmentation Combined with Word Embedding Technology. Aut. Control Comp. Sci. 54, 560–571 (2020). https://doi.org/10.3103/S0146411620060024
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0146411620060024