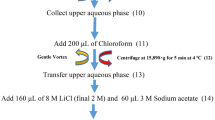
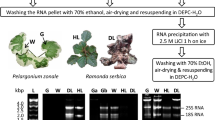
Abstract
Together with their undeniable role in the ecology of arid and semiarid ecosystems, Agave species are emerging as a model to dissect the relationships between crassulacean acid metabolism and high efficiency of light and water use, and as an energy crop for bioethanol production. Transcriptome resources from economically valuable Agaves species, such as Agave tequilana and A. salmiana, as well as hybrids for fibers, are now available, and multiple gene expression landscape analyses have been reported. Key components in molecular mechanisms underlying drought tolerance could be uncovered by analyzing gene expression patterns of roots. This study describes an efficient protocol for high-quality total RNA isolation from phenolic compounds-rich Agave roots. Our methodology involves suitable root handling and collecting in the field and using saving-time commercial kits available. RNA isolated from roots free of lignified out-layers and clean cortex showed high values of quality and integrity according to electrophoresis and microfluidics-based platform. Synthesis of long full-length cDNAs and PCR amplification tested the suitability for downstream applications of extracted RNA. The protocol was applied successfully to A. tequilana roots but can be used for other Agave species that also develop lignified epidermis/exodermis in roots.




Similar content being viewed by others
References
Abraham PE, Yin H, Borland AM et al (2016) Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat Plants 2:16178. https://doi.org/10.1038/nplants.2016.178
Abraham-Juárez MJ, Hernández-Cárdenas R, Santoyo-Villa JN et al (2015) Functionally different PIN proteins control auxin flux during bulbil development in Agave tequilana. J Exp Bot 66:3893–3905. https://doi.org/10.1093/jxb/erv191
Alemzadeh A, Fujie M, Usami S, Yamada T (2005) Isolation of high-quality RNA from high-phenolic tissues of eelgrass (Zostera marina L.) by keeping temperature low. Plant Mol Biol Rep 23:421. https://doi.org/10.1007/BF02788892
Avila-de Dios E, Gomez-Vargas AD, Damian-Santos ML, Simpson J (2015) New insights into plant glycoside hydrolase family 32 in Agave species. Front Plant Sci 6:594. https://doi.org/10.3389/fpls.2015.00594
Avila-de Dios E, Delaye L, Simpson J (2019) Transcriptome analysis of bolting in A. tequilana reveals roles for florigen, MADS, fructans and gibberellins. BMC Genomics 20:473. https://doi.org/10.1186/s12864-019-5808-9
Ávila-Fernández Á, Olvera-Carranza C, Rudiño-Piñera E et al (2007) Molecular characterization of sucrose: sucrose 1-fructosyltransferase (1-SST) from Agave tequilana Weber var. azul. Plant Sci 173:478–486. https://doi.org/10.1016/j.plantsci.2007.07.009
Batcho-Agossa A, Sarwar-Muhammad B, Tariq L et al (2019) Identification and characterisation of heat shock protein gene (HSP70) family and its expression in Agave sisalana under heat stress. J Hortic Sci Biotechnol 1–13:470–482. https://doi.org/10.1080/14620316.2019.1685412
Cervantes-Pérez SA, Espinal-Centeno A, Oropeza-Aburto A et al (2018) Transcriptional profiling of the CAM plant Agave salmiana reveals conservation of a genetic program for regeneration. Dev Biol 442:28–39. https://doi.org/10.1016/j.ydbio.2018.04.018
Corbin-Kendall R, Byrt-Caitlin S, Bauer S et al (2015) Prospecting for energy-rich renewable raw materials: agave leaf case study. PLoS ONE 10:e0135382. https://doi.org/10.1371/journal.pone.0135382
Cruz-Ramírez A, Calderón-Vázquez C, Herrera-Estrella L (2009) Effect of nutrient availability on root system development. Annual Plant Reviews Volume 37: Root Development 288–324. https://doi.org/10.1002/9781444310023.ch11.
Davis-Sarah C, Dohleman-Frank G, Long-Stephen P (2011) The global potential for Agave as a biofuel feedstock. Gcb Bioenergy. https://doi.org/10.1111/j.1757-1707.2010.01077.x
Deng G, Huang X, Xie L et al (2019) Identification and expression of SAUR Genes in the CAM Plant Agave. Genes 10:555. https://doi.org/10.3390/genes10070555
Dubrovsky JG (1997) Determinate primary-root growth in seedlings of Sonoran desert Cactaceae; its organization, cellular basis, and ecological significance. Planta 203:85–92. https://doi.org/10.1007/s00050168
Escamilla-Treviño LL (2012) Potential of plants from the genus Agave as bioenergy crops. BioEnergy Res 5:1–9. https://doi.org/10.1007/s12155-011-9159-x
Gao J, Yang F, Zhang S et al (2014) Expression of a hevein-like gene in transgenic Agave hybrid No. 11648 enhances tolerance against zebra stripe disease. Plant Cell Tissue Organ Cult 119:579–585. https://doi.org/10.1007/s11240-014-0557-6
Good-Avila SV, Souza V, Gaut BS, Eguiarte LE (2006) Timing and rate of speciation in Agave (Agavaceae). Proc Natl Acad Sci USA 103:9124–9129. https://doi.org/10.1073/pnas.0603312103
Graham GC (1993) A method for extraction of total RNA from Pinus radiata and other conifers. Plant Mol Biol Rep 11:32–37. https://doi.org/10.1007/BF02670557
Gross SM, Martin JA, Simpson J et al (2013) De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genomics 14:563. https://doi.org/10.1186/1471-2164-14-563
Guan L, Ma X, Zhou X et al (2019) An optimized method to obtain high-quality RNA from cassava storage root. 3 Biotech 9:118. https://doi.org/10.1007/s13205-019-1608-9
Handakumbura PP, Hazen SP (2012) Transcriptional regulation of grass secondary cell wall biosynthesis: playing catch-up with Arabidopsis thaliana. Front Plant Sci 3:74. https://doi.org/10.3389/fpls.2012.00074
Hernández-Castellano S, Garruña-Hernández R, Us-Camas R et al (2020) Agave angustifolia albino plantlets lose stomatal physiology function by changing the development of the stomatal complex due to a molecular disruption. Mol Genet Genomics. https://doi.org/10.1007/s00438-019-01643-y
Huang B, Nobel PS (1992) Hydraulic conductivity and anatomy for lateral roots of Agave deserti during root growth and drought-induced abscission. J Exp Bot 43:1441–1449. https://doi.org/10.1093/jxb/43.11.1441
Huang X, Xiao M, Xi J et al (2019a) De novo transcriptome assembly of Agave H11648 by Illumina sequencing and identification of cellulose synthase genes in Agave species. Genes 10:103. https://doi.org/10.3390/genes10020103
Huang X, Xie L, Gbokie T et al (2019b) Transcriptome dataset of leaf tissue in Agave H11648. Brown Univ Dig Addict Theory Appl 4:62. https://doi.org/10.3390/data4020062
Huded AKC, Jingade P, Mishra MK (2018) A rapid and efficient SDS-based RNA isolation protocol from different tissues of coffee. 3 Biotech 8:183. https://doi.org/10.1007/s13205-018-1209-z
Kadam PV, Yadav KN, Deoda RS et al (2012) Pharmacognostic and phytochemical studies on roots of Agave Americana (Agavaceae). Intern J Pharmacogn Phytochem Res 4:92–96
Kapp N, Barnes WJ, Richard TL, Anderson CT (2015) Imaging with the fluorogenic dye Basic Fuchsin reveals subcellular patterning and ecotype variation of lignification in Brachypodium distachyon. J Exp Bot 66:4295–4304. https://doi.org/10.1093/jxb/erv158
Martínez-Hernández A, Mena-Espino M, Herrera-Estrella AH et al (2010) Construcción de bibliotecas de ADNc y análisis de expresión génica por RT-PCR en agaves. Revista latinoamericana de química 38:21–44
McCahill IW, Hazen SP (2019) Regulation of cell wall thickening by a medley of mechanisms. Trends Plant Sci. https://doi.org/10.1016/j.tplants.2019.05.012
Niechayev NA, Jones AM, Rosenthal DM, Davis SC (2019) A model of environmental limitations on production of Agave americana L. grown as a biofuel crop in semi-arid regions. J Exp Bot 70:6549–6559. https://doi.org/10.1093/jxb/ery383
Nobel PS (1976) Water relations and photosynthesis of a desert CAM plant, Agave deserti. Plant Physiol 58:576–582. https://doi.org/10.1104/pp.58.4.576
North GB, Nobel PS (1991) Changes in hydraulic conductivity and anatomy caused by drying and rewetting roots of Agave deserti (Agavaceae). Am J Bot 78:906–915. https://doi.org/10.1002/j.1537-2197.1991.tb14494.x
North GB, Nobel PS (1995) Hydraulic conductivity of concentric root tissues of Agave deserti Engelm. under wet and drying conditions. New Phytol 130:47–57. https://doi.org/10.1111/j.1469-8137.1995.tb01813.x
Rubio-Piña JA, Zapata-Pérez O (2011) Isolation of total RNA from tissues rich in polyphenols and polysaccharides of mangrove plants. Electron J Biotechnol
Sara HC, René GH, Rosa UC et al (2020) Agave angustifolia albino plantlets lose stomatal physiology function by changing the development of the stomatal complex due to a molecular disruption. Mol Genet Genomics 295:787–805. https://doi.org/10.1007/s00438-019-01643-y
Sarwar MB, Ahmad Z, Rashid B et al (2019) De novo assembly of Agave sisalana transcriptome in response to drought stress provides insight into the tolerance mechanisms. Sci Rep 9:396. https://doi.org/10.1038/s41598-018-35891-6
Sierra-Gómez Y, Rodríguez-Hernández A, Cano-Sánchez P et al (2019) A biophysical and structural study of two chitinases from Agave tequilana and their potential role as defense proteins. FEBS J 286:4778–4796. https://doi.org/10.1111/febs.14993
Acknowledgements
This work was funded by CONACyT (Grant 1049) and Colpos. Luis F. Maceda-Lopez is grateful for a M.Sc. fellowship from CONACyT. Eleazar Garcia-Hernández and Dalia C. Moran Velazquez were supported by an undergraduate scholarship from CONACyT (Grant 1049). Authors especially grateful to Victor H. Fernandez-Carrillo, manager of Santa Genoveva farm, for supporting during Agave samples collecting.
Author information
Authors and Affiliations
Contributions
LFML, JLVA, DCMV, and FAC designed and performed the RNA extraction, quantification, and quality RNA analysis. JLVA, EGH, LRL, DHD, and MACV performed RT-PCR analysis. Lignin deposition and SEM analysis were carried out by LFML, SBAC, LRL, and FAC. Data mining was performed by LFML, EAD, JS, LT, ILR, EGC, and FAC. LFML, LT, EGC, ILR, JS, and FAC discussed results. LFML and FAC conceived and designed the research and wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Ethical approval
Each of the authors confirms that this manuscript is original, has not been previously published, and is not currently under consideration by any other journal. Additionally, all of the authors have approved the contents of this paper and have agreed to the 3 Biotech’s submission policies.
Rights and permissions
About this article
Cite this article
Maceda-López, L.F., Villalpando-Aguilar, J.L., García-Hernández, E. et al. Improved method for isolation of high-quality total RNA from Agave tequilana Weber roots. 3 Biotech 11, 75 (2021). https://doi.org/10.1007/s13205-020-02620-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02620-8