Stress Granules in the Post-transcriptional Regulation of Immune Cells
- 1Cancer Research Center of Toulouse (CRCT), INSERM UMR 1037, CNRS ERL 5294, Toulouse, France
- 2Université Toulouse III Paul Sabatier, Toulouse, France
- 3Institut Universitaire du Cancer de Toulouse-Oncopole, Toulouse, France
- 4Département de Pathologie, Centre Hospitalier Universitaire (CHU) de Toulouse, Toulouse, France
Immune cell activation triggers transcriptional and translational programs eliciting cellular processes, such as differentiation or proliferation, essential for an efficient immune response. These dynamic processes require an intricate orchestration of regulatory mechanisms to control the precise spatiotemporal expression of proteins. Post-transcriptional regulation ensures the control of messenger RNA metabolism and appropriate translation. Among these post-transcriptional regulatory mechanisms, stress granules participate in the control of protein synthesis. Stress granules are ribonucleoprotein complexes that form upon stress, typically under control of the integrated stress response. Such structures assemble upon stimulation of immune cells where they control selective translational programs ensuring the establishment of accurate effector functions. In this review, we summarize the current knowledge about post-transcriptional regulation in immune cells and highlight the role of stress sensors and stress granules in such regulation.
Introduction
Immune cell compartment is composed of two main cell types, the myeloid cells, such as macrophages or dendritic cells (DC), and the lymphoid cells, namely B and T lymphocytes. Macrophages and DC are key components of the innate immunity. They sense pathogenic threat and trigger adaptive immune response by presenting antigenic peptides to lymphoid cells. Upon antigen (Ag) recognition by their cell surface receptors [surface immunoglobulin (Ig) and T-cell receptor (TCR) in B and T cells, respectively], the lymphocytes become fully functional.
For all these immune cells, Ag stimulation triggers transcriptional and translational programs of differentiation, proliferation, and effector functions. Hence, the dynamics of immune response requires a tight control in gene expression, which can be achieved through post-transcriptional regulation. One important aspect of this regulation implies a decorrelation of transcriptional and translational activities for a given messenger RNA (mRNA) (de Sousa Abreu et al., 2009; Schwanhäusser et al., 2011; Buccitelli and Selbach, 2020). This regulation is mediated by multiple RNA-binding proteins (RBP) that perform their function on cis-acting elements typically lying on the 5′ and 3′ untranslated region (UTR) of mRNA (Gehring et al., 2017; Leppek et al., 2017; Mayr, 2017).
Actually, Ag stimulation and the ensuing cellular processes represent and are sensed as a physiological stress by immune cells. A general sensor of cellular stress is the integrated stress response (ISR), an evolutionary conserved signaling pathway that helps cells to adapt to environmental stress (Brostrom et al., 1996; Harding et al., 2000, 2003; Scheuner et al., 2001; Pakos-Zebrucka et al., 2016; Costa-Mattioli and Walter, 2020). At the center of ISR lies the eukaryotic initiation factor 2 (eIF2), which can be phosphorylated on its α subunit by four different kinases (Taniuchi et al., 2016), namely heme-regulated inhibitor (HRI or EIF2AK1) (Chen et al., 1991), protein kinase R (PKR or EIF2AK2) (Kostura and Mathews, 1989), PKR-like endoplasmic reticulum (PERK or EIF2AK3) (Harding et al., 1999), and general control non-repressible 2 (GCN2 or EIF2AK4) (Dever et al., 1992). Once phosphorylated, eIF2α prevents the formation of the ternary complex (eIF2:GTP:methionyl-initiator-tRNA) and consequently blocks translation initiation. Hence, activation of ISR globally inhibits translation while selectively preserving the expression of genes, including the transcription factor ATF4, to facilitate cell recovery (Wong et al., 1993; Brostrom et al., 1996; Harding et al., 2000; Lu et al., 2004; Schneider et al., 2020).
Importantly, this inhibition of translation caused by eIF2α phosphorylation seeds the formation of ribonucleoprotein complexes called stress granules (SG) (Kedersha et al., 2002; Panas et al., 2016; Protter and Parker, 2016). These structures sequester RNA upon stress and maintain a translation arrest until stress releases. SG can also form independently of eIF2α phosphorylation due to modifications in the eIF4F complex (comprising eIF4E, eIF4A, and eIF4G) (Emara et al., 2012; Fujimura et al., 2012; Panas et al., 2016). This complex is regulated by the mechanistic target of rapamycin (mTOR) kinase, acting as a metabolic sensor and regulator of translation (Ma and Blenis, 2009). When mTOR is inactivated, the eIF4E-binding protein 1 (4E-BP1) is no longer phosphorylated and prevents the formation of the eIF4F complex, which inhibits translation initiation and allows SG formation.
Understanding these post-transcriptional regulations and their mechanisms in immune cells is necessary and could help to improve treatments of autoimmunity and cancer. Here, we review the post-transcriptional regulation that are linked with SG in immune cells (Figure 1). Notably, this review will outline their involvement during T-cell activation, their control of cytokine expression, the influence of environmental cues, and how such mechanisms also control the expression of immune checkpoints (IC) in activated T lymphocytes. The following paragraphs will review the concurrent findings about the stress-dependent regulation of Ig diversification in B lymphocytes, of Ag presentation in DC, and finally of cytokine expression in myeloid cells.
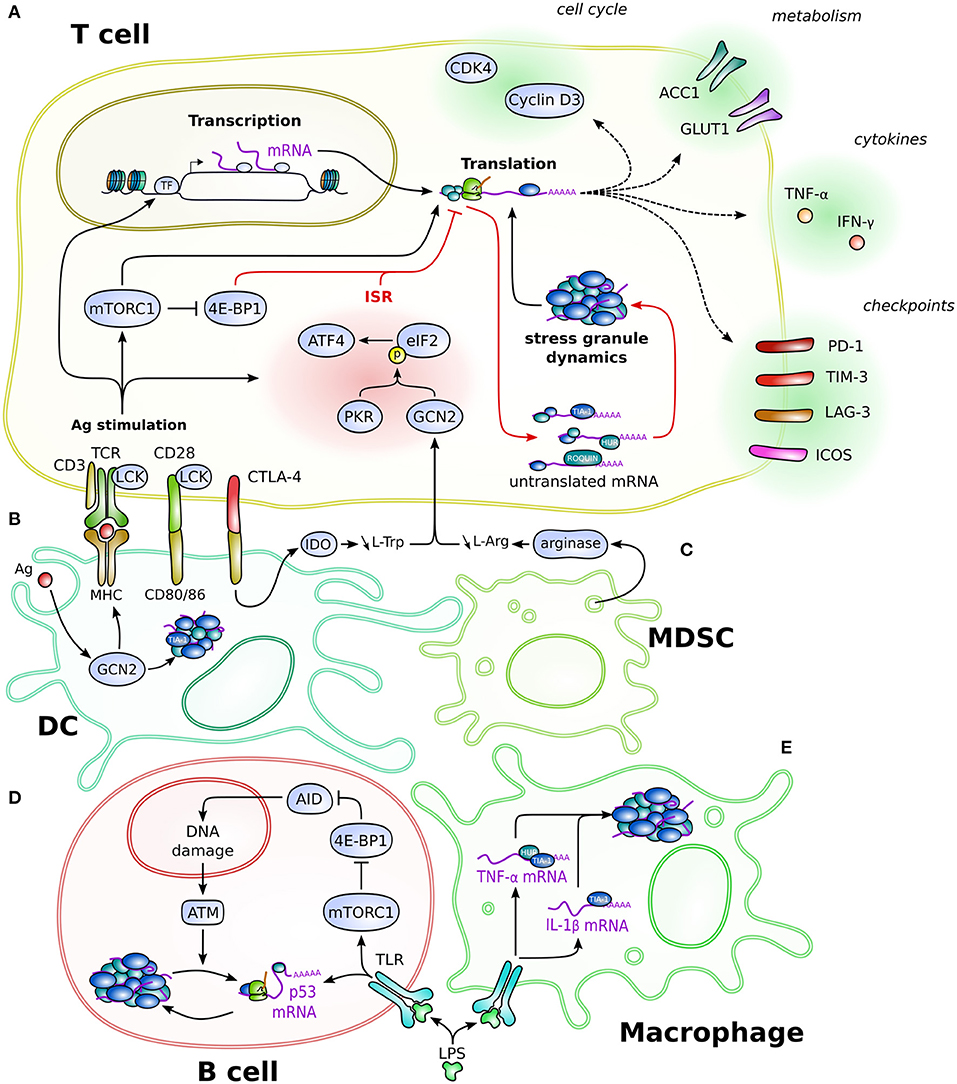
Figure 1. Schematic overview of the SG-based post-transcriptional regulation in immune cells. Depicted is the regulation in and connection between T cells (A), DC (B), MDSC (C), B cells (D), and macrophages (E). Post-transcriptional mechanisms regulate different aspects of cellular biology, such as cell cycle, cytokine production, metabolism, or regulatory proteins like immune checkpoints, eventually impacting on cellular processes such as proliferation, differentiation, and effector functions. Red arrows specify the direct effector leading to translation inhibition and formation of stress granules. Dotted line arrows represent the product of translation that occurs once the mRNA is released from the post-transcriptional regulation. Cell-type-specific regulation is detailed in the text. Cells are not drawn at scale.
General Post-Transcriptional Regulation During T-Lymphocyte Activation
Prior to antigenic stimulation, T cells are actively maintained in a quiescent state due to post-transcriptional processes allowing high turnover and rapid degradation of RNA while maintaining a substantial amount of functional yet inactive ribosomal proteins (Ricciardi et al., 2018; Hwang et al., 2020; Wolf et al., 2020). Genome-wide “omics” analyses of transcriptome, translatome, and proteome during T-cell activation and differentiation pointed out a major role of post-transcriptional regulatory mechanisms (Hukelmann et al., 2016; Araki et al., 2017; Tan et al., 2017a,b; Howden et al., 2019) (Table 1). During these transitions, the regulation of ribosomal subunits expression and the activity of the mTOR complex 1 (mTORC1) signaling pathway are essential for shaping the proteomic landscape of T lymphocytes.
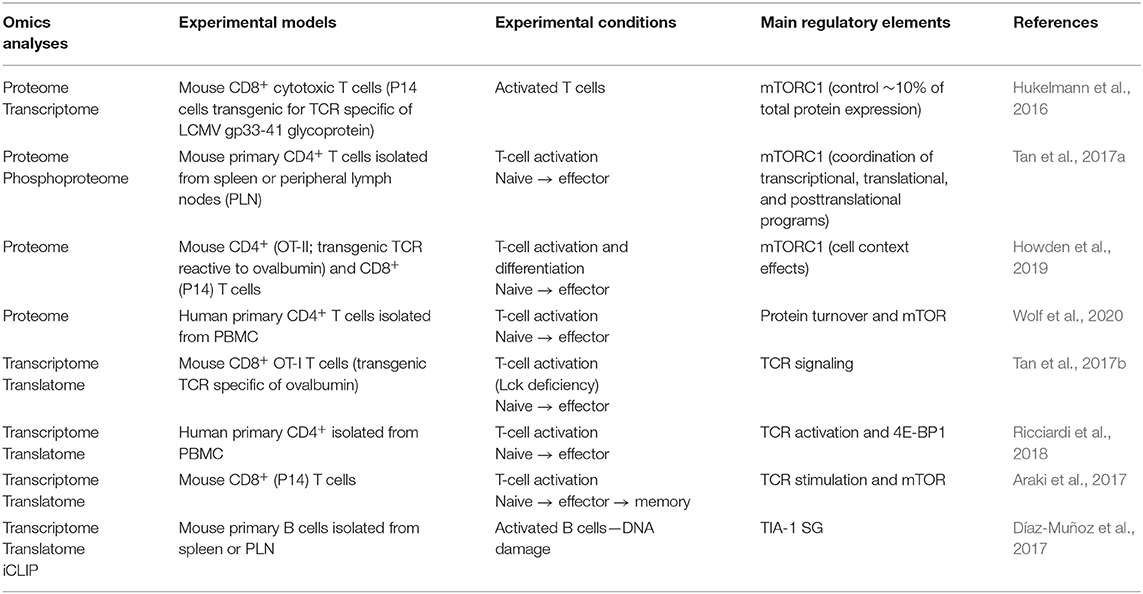
Table 1. Omics approaches suggesting post-transcriptional and translational regulation in immune cells.
These features primarily depend on the TCR stimulation and its underlying signaling. This has been assessed by deleting Lck, one essential component of the TCR-mediated signal transduction complex, in CD8+ T cells (Tan et al., 2017b) (Figure 1A). In comparison with wild-type cells, Lck-deficient counterparts displayed a reduced mTOR signaling activity, reflected by the accumulation of unphosphorylated 4E-BP1, and failed to properly induce ribosome biogenesis. Consequently, overall protein synthesis was limited, thereby impairing the proliferative ability. The analysis of global RNA expression and their association with polysomes, which reflect the translation efficiency, in Lck-deficient T cells confirmed that one fourth of all mRNA were post-transcriptionally regulated (Tan et al., 2017b).
One important change that comes with immune cell activation and differentiation is metabolic reprogramming (Chapman et al., 2020). Metabolic analyses identified the clustering of T-cell subsets (Ricciardi et al., 2018) and defined the metabolic reprogramming triggered by T-cell activation (Ricciardi et al., 2018; Wolf et al., 2020) (Figure 1A). Interestingly, in naive CD4+ T cells, GLUT1 and ACC1 proteins, which, respectively, control glucose entry and fatty acid synthesis, are absent, while their corresponding mRNA are abundant and whose fraction is already associated with polysomes (Ricciardi et al., 2018). At this stage, 4E-BP1 is expressed and outnumbers eIF4E, and mTOR activity is low, indicating a control at the translation level (Ricciardi et al., 2018; Wolf et al., 2020). Interestingly, while TCR stimulation, which induces mTORC1 activation, promotes GLUT1 and ACC1 protein expression, there is still no correlation between the kinetics of appearance of mRNA vs. protein and neither with their association with polysomes (Ricciardi et al., 2018). This control of translation through the modulation of the eIF4F complex activity was further explored during T-cell polarization. Because mTORC1 has a dual translational regulatory activity, as repressor and inductor, this modulation could be necessary to selectively adjust translational programs to lineage identity (Hukelmann et al., 2016; Howden et al., 2019). In CD4+ T lymphocytes stimulated under T helper (Th) 17 polarization conditions, inhibition of eIF4E limits Th17 differentiation to promote Foxp3-expressing regulatory T-cell (Treg) development (Ricciardi et al., 2018). In agreement, genome-wide analysis of polysome-associated mRNA revealed that Treg have distinct translational abilities compared with non-Treg (Foxp3−) cells, mainly based on eIF4E expression, which was not detected in Treg (Bjur et al., 2013).
Thus, TCR stimulation controls selective translational activity through post-transcriptional mechanisms that maintain mRNA in complexes with repressive translational configuration and/or control their delivery to the polysomes.
Control of Cytokine Expression by Stress-Dependent Mechanisms in T Lymphocytes
Analysis of translation regulation among the naive, effector (early and late, based on their proliferation status) and memory CD8+ T cells in mice during infection with the lymphocytic choriomeningitis virus (LCMV) clearly indicated a different translational ability during the activation and differentiation processes (Araki et al., 2017). Typically, Ifng mRNA levels increased after infection in early and late effector cells, while IFN-γ proteins peaked in early effectors and decrease in late effectors, correlating with the localization of the mRNA within the polysome fractions. In contrast, Granzyme B, a cytotoxic effector, is expressed in early effector cells but not in naive cells, which correlates with the level of mRNA expression and its association with polysomes. However, while the level of mRNA and the association to polysomes were similar in late effector and memory cells, these cells gradually lose the protein expression (Araki et al., 2017).
Thus, active post-transcriptional mechanisms control mRNA translation during T-cell activation by regulating the mRNA delivery to the polysomes and/or reconfiguring protein complexes controlling mRNA translation. Such modulation of translational regulatory complexes is controlled by stress sensor kinases, which are upregulated in stimulated T cells (Howden et al., 2019) (Figure 1A). In CD4+ T cells, the GCN2–eIF2α-ATF4 pathway favors Th1 over Th17 differentiation and is required for IFN-γ expression (Sundrud et al., 2009; Ravindran et al., 2016; Yang et al., 2018). In addition, it was suggested that IFNG mRNA regulates its own expression by activating PKR through a peculiar structure, called pseudoknot, in its 5′UTR (Ben-Asouli et al., 2002; Cohen-Chalamish et al., 2009). Binding of PKR on this pseudoknot results in eIF2α phosphorylation, which inhibits IFNG mRNA translation. Likewise, TNFA mRNA contains such pseudoknot in its 3′UTR (Osman et al., 1999). Recognition of this structure by PKR also leads to eIF2α phosphorylation. However, in this particular case, this signaling is required for efficient splicing of TNFA mRNA, suggesting an additional regulatory function for this pathway (Namer et al., 2017).
Under persistent antigen stimulation, mRNA encoding ribosomal proteins are still associated with active polysome fractions, indicating that stimulation maintains translation activity (Araki et al., 2017). In addition to ribosomal proteins, Il4 mRNA is also relocated to active polysome fractions upon TCR re-engagement of primed CD4+ T lymphocytes under Th2 conditions (Scheu et al., 2006). Interestingly, T-cell priming triggers ISR, reflected by eIF2α phosphorylation, and SG, reflected by TIA-1-positive granule formation. Upon re-stimulation, the eIF2α phosphorylation decreases, which correlates with the increases of the translation activity. Moreover, deletion of TIA-1 bypasses the need of re-stimulation to produce the IL-4 protein, suggesting that the post-transcriptional regulation operating on Il4 mRNA is mediated by the SG. However, TIA-1-positive granules persist upon re-stimulation, raising the question if Il4 mRNA is directly regulated by SG, but anyhow indicating that SG is required throughout T-cell activation. Given the general variation in translation activity relative to T-cell activation, it is likely that the SG-dependent regulation is not restricted to cytokines (Scheu et al., 2006; Araki et al., 2017).
Environmental Cues Influencing Stress-Dependent Regulation in T Lymphocytes
While antigenic stimulation induces important reorganization in T cells, other environmental signals can significantly influence T-cell biological functions (Figures 1A–C). Plasmacytoid DC from tumor-draining lymph nodes express indoleamine 2,3-dioxygenase (IDO), an enzyme involved in tryptophan starvation. IDO expression by plasmacytoid DC induces a GCN2-dependent proliferative arrest in T cells (Munn et al., 2005) (Figures 1A,B). Similar issues are induced by L-arginine starvation (Rodriguez et al., 2007), which can be caused by myeloid-derived suppressor cells that express high amounts of arginase (Figures 1A,C). T cell activated in such conditions failed to proliferate due to a lack of expression of cyclin D3 and cyclin-dependent kinase 4 (cdk4), which regulate cell cycle progression. Furthermore, the RBP HuR interacts with the 3′UTR of the cyclin D3 mRNA to stabilize it (Rodriguez et al., 2010). Upon activation under L-arg deprivation, GCN2 is activated, which decreases the expression of HuR protein, but not the mRNA. Consequently, cyclin D3 mRNA is no longer stabilized and does not proceed to translation. In addition to cyclin D3, a global translation inhibition due to L-arg deprivation was observed, suggesting that this regulatory mechanism is general (Rodriguez et al., 2010). Moreover, GCN2 activation is involved in tolerogenic mechanisms (Cobbold et al., 2009). While the proliferative arrest due to GCN2 activation promotes T-cell survival, this phenotype also depends on the mTOR pathway, as amino-acid (AA) starvation decreases 4E-BP1 phosphorylation. Along with GCN2, mTOR signaling provides a positive feedback for the generation of Treg. In addition, Treg promote T-cell proliferation arrest through a CTLA-4-dependent signaling that stimulates expression of AA-consuming enzymes, such as IDO, by DC (Cobbold et al., 2009) (Figures 1A,B).
The complexity of pathway interconnection could explain the role of GCN2 in controlling proliferation and trafficking of T lymphocytes upon stimulation (Van de Velde et al., 2016). Expression of ATF4 was partially dependent on GCN2 activity under AA deprivation and oxidizing environment in activated Th1-polarized CD4+ T cells (Yang et al., 2018). This ATF4 pathway could regulate mTORC1 activity under oxidative conditions and drive the metabolic reprogramming upon CD4+ T-cell stimulation (Yang et al., 2018), to allow proliferation and T-cell function (Ricciardi et al., 2018; Wolf et al., 2020).
Control of Immune Checkpoint Expression by Stress-Dependent Mechanisms in T Lymphocytes
IC receptors play essential regulatory functions in immune cells and represent therapeutic targets for autoimmune diseases and cancers (Zhang and Vignali, 2016; Ribas and Wolchok, 2018). Among the different levels of regulation controlling IC expression, SG-based post-transcriptional mechanisms are implicated and represent important new therapeutic targets (Curdy et al., 2019; Gao et al., 2019) (Figure 1A).
In human CD4+ and CD8+ T lymphocytes, stimulation of resting cells induces the expression of SG markers and the formation of SG, which selectively control inhibitory IC expression (Franchini et al., 2019). This SG-dependent regulation relies on microtubules and the molecular motor kinesin1, with the 3′UTR of mRNA being essential, notably for PD-1 expression. When T lymphocytes are treated with microtubule-targeting drugs, the translation of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and BTLA is inhibited (Franchini et al., 2019).
ROQUIN, a protein that plays an important role in preventing autoimmune diseases, is commonly found in SG. ROQUIN limits the inducible T-cell co-stimulator (ICOS) expression in CD4+ T cells and was found to bind directly to the 3′UTR of the ICOS mRNA, and both co-localize in SG (Yu D. et al., 2007; Athanasopoulos et al., 2010; Glasmacher et al., 2010). However, they are also found in processing bodies, another ribonucleoprotein complex involved in RNA degradation (Yu D. et al., 2007; Glasmacher et al., 2010), suggesting that in T lymphocytes, both types of granules are interconnected, as in other cell types (Protter and Parker, 2016). Moreover, the 3′UTR-dependent inhibitory regulation of ROQUIN was also observed with OX40, another stimulatory IC, suggesting a broader action of the SG-dependent regulation on stimulatory IC (Vogel et al., 2013).
Stress-Dependent Regulation of Immunoglobulin Diversity in B Lymphocytes
Ig and TCR assembly relies on a series of genomic rearrangements that are intimately linked with DNA damage repair. In addition, mature B cells stimulated in germinal centers trigger the expression of the activation-induced deaminase (AID), which initiates class switch recombination and somatic hypermutation to increase Ig affinity and specificity toward the Ag (Franchini and Petersen-Mahrt, 2014). Following stimulation of primary murine B cells, TIA-1-positive SG are formed and sequester a substantial amount of mRNA, mainly through binding to their 3′UTR (Díaz-Muñoz et al., 2017) (Figure 1D). Among those mRNA lies p53, a key factor of DNA damage response. Upon etoposide treatment, an inhibitor of topoisomerase II that induces DNA double strand breaks used to mimic AID-induced DNA damage, ATM kinase signaling is triggered, releasing p53 mRNA from SG and relocating it to active polysomes for translation, thereby promoting DNA damage repair (Díaz-Muñoz et al., 2017). Intriguingly, AID expression is controlled by 4E-BP1, and mTOR inhibition with rapamycin decreases AID protein but not its transcript (Chiu et al., 2019) (Figure 1D). Therefore, a sequential SG-dependent post-transcriptional regulation most likely underlies B-cell differentiation.
Stress-Dependent Regulation of Antigen Presentation in Dendritic Cells
DC are able to sense pathogenic threat and to initiate adaptive immune response through Ag presentation (Pulendran, 2015). Following inoculation of the yellow fever virus vaccine, the kinase GCN2 is activated in DC (Ravindran et al., 2014) (Figure 1B). This activation strongly correlates with the subsequent T-cell response. Mechanistically, while GCN2 deficiency does not impair the secretion of inflammatory cytokines, it reduces autophagy-mediated antigen presentation and, consequently, impairs T-cell priming (Ravindran et al., 2014). In addition, stimulation of DC by vaccination induces the formation of TIA-1-positive SG (Ravindran et al., 2014). Although SG were shown to be crucial during innate antiviral immune response (McCormick and Khaperskyy, 2017), the real function of TIA-1 granules in that case was not established.
Control of Cytokine Expression by Stress-Dependent Mechanisms in Myeloid Cells
Early studies revealed the importance of post-transcriptional regulation in the control of cytokine expression (Ivanov and Anderson, 2013). Upon stimulation, macrophages isolated from Tia-1-deficient mice have an increased proportion of Tnfa mRNA associated with polysomes and, consequently, overexpress TNF-α protein, which suggests a TIA-1-dependent control of mRNA translation (Piecyk et al., 2000). Interestingly, TIA-1-deficient lymphocytes do not overexpress TNF-α upon stimulation, indicating that this TIA-1-dependent regulation is cell-type-specific (Saito et al., 2001). Along with TIA-1, the RBP HuR and the protein steroid receptor coactivator (SRC-3) co-localize in SG following arsenite treatment, and they silence Tnfa mRNA translation in LPS-stimulated macrophages (Katsanou et al., 2005; Yu C. et al., 2007) (Figure 1E).
Pharmacological activation of the ISR pathway GCN2–eIF2α in LPS-stimulated macrophages controls IL-1β production by a mechanism involving TIA-1-positive SG (Battu et al., 2018; Naz et al., 2019) (Figure 1E). Likewise, deletion of Gcn2 or Atf4 in myeloid cells abrogates IL-1β expression (Ravindran et al., 2016; Yang et al., 2018). This influences CD4+ T-lymphocyte polarization in preventing Th17 differentiation and, therefore, synergizes with the intrinsic role of this ISR pathway in CD4+ T lymphocytes to favor Th1 differentiation (Sundrud et al., 2009; Ravindran et al., 2016; Yang et al., 2018). Modulation of this pathway in mice models allowed to reduce inflammation and protect against autoimmune disorders (Sundrud et al., 2009; Ravindran et al., 2016).
Conclusion
External stimuli sensed by immune cells trigger specific functional programs that allow the completion of immune response and pathogenic clearance. It is now clear that the rapid variation in gene expression in stimulated immune cells is not only controlled at the transcriptional level but also implies post-transcriptional regulation, with SG emerging as a key regulator. The immune response relies on an intricate connection between the integration of environmental signals and the communication between cells, which leads to the specific development and differentiation of cell types. Although each cell type aims at providing a specific function, they all use similar regulatory mechanisms to control their protein expression. During immune cell activation, the ISR pathway, through the kinases that all converge on eIF2α, as well as the mTOR/4E-BP1 pathway, is activated. These pathways aim at SG formation for the control of mRNA translation.
The immune response, the underlying stress-dependent signaling pathways, and the biochemical properties of SG are all very dynamic. Such dynamic features would confer a greater versatility for immune cells to properly shape immune response. The intricate signaling network emphasizes the complexity of the control of immune response and suggests that each signal and resulting SG provide different regulatory outcomes. Whether each signaling pathway has regulatory functions in the different immune cell types, and to which extent, is still an open question. It will be therefore important to associate the dynamics of immune response with the activation of the different stress-dependent signaling pathways. Integrative single-cell molecular analyses are required for deciphering such interplay and to assign it within the immune cell population during the different cellular processes they undergo.
Proteome and transcriptome modulation ensuing from the integration of these distinct signaling pathways could influence SG constitution and function and dictate the dynamics of mRNA exchange within SG. It is now clear that the proteomic composition of SG is heterogeneous and depends on the nature of the stress and the cell type (Markmiller et al., 2018). Although TIA-1 was mainly used to identify SG in immune cells, one important aspect to resolve is whether different types of SG are formed in immune cells. Moreover, in addition to the primary stimulus, the influence of environmental factors, such as AA availability or cytokines, on SG formation and composition needs to be determined. This could provide targets to manipulate mRNA translation and will clarify the degree of heterogeneity in immune SG.
Besides SG proteomics, the evolution of the mRNA content depending on the cellular state of immune cells also needs to be addressed. While mRNA UTR appears important for SG targeting, they are themselves subjected to regulatory mechanisms (Mayr, 2016; Leppek et al., 2017). Cellular stresses modify 3′UTR mRNA features (Mizrahi et al., 2018; Zheng et al., 2018), which can eventually impact on RBP binding, such as TIA-1, and SG formation (Zheng et al., 2018). Similar 3′UTR modifications were observed in activated and proliferating lymphocytes (Sandberg et al., 2008; Gruber et al., 2014). While transcriptomic analyses are required to determine the mRNA content of SG and evaluate the regulatory function of SG, additional analyses that precisely characterize the mRNA features will be necessary.
The complex post-transcriptional regulation performed by SG warrants future studies to determine their role in the different functional programs of immune cells. Although proteomic and transcriptomic analyses of their SG will shed light on this regulation, another challenge will be to apply these studies to primary immune cells. To overcome these limitations, improvement and adaptation of single-cell analyses might turn out to be decisive for integrative elucidation of such regulation in immune cells. Their deciphering has the potential to lead to the development of novel therapeutic approaches for autoimmune disorders, viral infections, and cancer.
Author Contributions
NC and D-MF conceptualized this review, decided on the content, and wrote the manuscript. NC and D-MF prepared the figures. OL, SC, CL, and J-JF revised this review. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work.
Funding
This work was supported by institutional funding from the INSERM, the Université de Toulouse 3, and the CNRS and by specific grants from the Laboratoire d'Excellence TouCan, Roche (imCORE), Ligue Contre Le Cancer, Cancéropôle Grand Sud-Ouest, and the Fondation ARC Pour la Recherche sur le Cancer (Équipe Labellisée) (N° PGA1 RF20190208691).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely apologize to colleagues whose work could not be included in this review owing to space limitations. We thank Anne Quillet-Mary for helpful discussions and critical reading of the manuscript.
References
Araki, K., Morita, M., Bederman, A. G., Konieczny, B. T., Kissick, H. T., Sonenberg, N., et al. (2017). Translation is actively regulated during the differentiation of CD8+ effector T cells. Nat. Immunol. 18, 1046–1057. doi: 10.1038/ni.3795
Athanasopoulos, V., Barker, A., Yu, D., Tan, A. H.-M., Srivastava, M., Contreras, N., et al. (2010). The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs: Roquin localizes to stress granules and binds RNA. FEBS J. 277, 2109–2127. doi: 10.1111/j.1742-4658.2010.07628.x
Battu, S., Afroz, S., Giddaluru, J., Naz, S., Huang, W., Khumukcham, S. S., et al. (2018). Amino acid starvation sensing dampens IL-1β production by activating riboclustering and autophagy. PLoS Biol. 16:e2005317. doi: 10.1371/journal.pbio.2005317
Ben-Asouli, Y., Banai, Y., Pel-Or, Y., Shir, A., and Kaempfer, R. (2002). Human interferon-γ mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell 108, 221–232. doi: 10.1016/S0092-8674(02)00616-5
Bjur, E., Larsson, O., Yurchenko, E., Zheng, L., Gandin, V., Topisirovic, I., et al. (2013). Distinct translational control in CD4+ T cell subsets. PLoS Genet. 9:e1003494. doi: 10.1371/journal.pgen.1003494
Brostrom, C. O., Prostko, C. R., Kaufman, R. J., and Brostrom, M. A. (1996). Inhibition of translational initiation by activators of the glucose-regulated stress protein and heat shock protein stress response systems: role of the interferon-inducible double-stranded RNA-activated eukaryotic initiation factor 2α kinase. J. Biol. Chem. 271, 24995–25002. doi: 10.1074/jbc.271.40.24995
Buccitelli, C., and Selbach, M. (2020). mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 21, 630–644. doi: 10.1038/s41576-020-0258-4
Chapman, N. M., Boothby, M. R., and Chi, H. (2020). Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70. doi: 10.1038/s41577-019-0203-y
Chen, J. J., Throop, M. S., Gehrke, L., Kuo, I., Pal, J. K., Brodsky, M., et al. (1991). Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF-2 alpha) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2 alpha kinase. Proc. Natl. Acad. Sci. U.S.A. 88, 7729–7733. doi: 10.1073/pnas.88.17.7729
Chiu, H., Jackson, L. V., Oh, K. I., Mai, A., Ronai, Z. A., Ruggero, D., et al. (2019). The mTORC1/4E-BP/eIF4E axis promotes antibody class switching in B lymphocytes. J. Immunol. 202, 579–590. doi: 10.4049/jimmunol.1800602
Cobbold, S. P., Adams, E., Farquhar, C. A., Nolan, K. F., Howie, D., Lui, K. O., et al. (2009). Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc. Natl. Acad. Sci. 106, 12055–12060. doi: 10.1073/pnas.0903919106
Cohen-Chalamish, S., Hasson, A., Weinberg, D., Namer, L. S., Banai, Y., Osman, F., et al. (2009). Dynamic refolding of IFN-γ mRNA enables it to function as PKR activator and translation template. Nat. Chem. Biol. 5, 896–903. doi: 10.1038/nchembio.234
Costa-Mattioli, M., and Walter, P. (2020). The integrated stress response: from mechanism to disease. Science 368:eaat5314. doi: 10.1126/science.aat5314
Curdy, N., Lanvin, O., Laurent, C., Fournié, J.-J., and Franchini, D.-M. (2019). Regulatory mechanisms of inhibitory immune checkpoint receptors expression. Trends Cell Biol. 29, 777–790. doi: 10.1016/j.tcb.2019.07.002
de Sousa Abreu, R., Penalva, L. O., Marcotte, E. M., and Vogel, C. (2009). Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526. doi: 10.1039/b908315d
Dever, T. E., Feng, L., Wek, R. C., Cigan, A. M., Donahue, T. F., and Hinnebusch, A. G. (1992). Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68, 585–596. doi: 10.1016/0092-8674(92)90193-G
Díaz-Muñoz, M. D., Kiselev, V. Y., Le Novère, N., Curk, T., Ule, J., and Turner, M. (2017). Tia1 dependent regulation of mRNA subcellular location and translation controls p53 expression in B cells. Nat. Commun. 8:530. doi: 10.1038/s41467-017-00454-2
Emara, M. M., Fujimura, K., Sciaranghella, D., Ivanova, V., Ivanov, P., and Anderson, P. (2012). Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 423, 763–769. doi: 10.1016/j.bbrc.2012.06.033
Franchini, D.-M., Lanvin, O., Tosolini, M., Patras de Campaigno, E., Cammas, A., Péricart, S., et al. (2019). Microtubule-driven stress granule dynamics regulate inhibitory immune checkpoint expression in T cells. Cell Rep 26, 94–107.e7. doi: 10.1016/j.celrep.2018.12.014
Franchini, D.-M., and Petersen-Mahrt, S. K. (2014). AID and APOBEC deaminases: balancing DNA damage in epigenetics and immunity. Epigenomics 6, 427–443. doi: 10.2217/epi.14.35
Fujimura, K., Sasaki, A. T., and Anderson, P. (2012). Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 40, 8099–8110. doi: 10.1093/nar/gks566
Gao, X., Jiang, L., Gong, Y., Chen, X., Ying, M., Zhu, H., et al. (2019). Stress granule: a promising target for cancer treatment. Br. J. Pharmacol. 176, 4421–4433. doi: 10.1111/bph.14790
Gehring, N. H., Wahle, E., and Fischer, U. (2017). Deciphering the mRNP code: RNA-bound determinants of post-transcriptional gene regulation. Trends Biochem. Sci. 42, 369–382. doi: 10.1016/j.tibs.2017.02.004
Glasmacher, E., Hoefig, K. P., Vogel, K. U., Rath, N., Du, L., Wolf, C., et al. (2010). Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat. Immunol. 11, 725–733. doi: 10.1038/ni.1902
Gruber, A. R., Martin, G., Müller, P., Schmidt, A., Gruber, A. J., Gumienny, R., et al. (2014). Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat. Commun. 5:5465. doi: 10.1038/ncomms6465
Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., et al. (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. doi: 10.1016/S1097-2765(00)00108-8
Harding, H. P., Zhang, Y., and Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274. doi: 10.1038/16729
Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633. doi: 10.1016/S1097-2765(03)00105-9
Howden, A. J. M., Hukelmann, J. L., Brenes, A., Spinelli, L., Sinclair, L. V., Lamond, A. I., et al. (2019). Quantitative analysis of T cell proteomes and environmental sensors during T cell differentiation. Nat. Immunol. 20, 1542–1554. doi: 10.1038/s41590-019-0495-x
Hukelmann, J. L., Anderson, K. E., Sinclair, L. V., Grzes, K. M., Murillo, A. B., Hawkins, P. T., et al. (2016). The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat. Immunol. 17, 104–112. doi: 10.1038/ni.3314
Hwang, S. S., Lim, J., Yu, Z., Kong, P., Sefik, E., Xu, H., et al. (2020). mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 367, 1255–1260. doi: 10.1126/science.aax0194
Ivanov, P., and Anderson, P. (2013). Post-transcriptional regulatory networks in immunity. Immunol. Rev. 253, 253–272. doi: 10.1111/imr.12051
Katsanou, V., Papadaki, O., Milatos, S., Blackshear, P. J., Anderson, P., Kollias, G., et al. (2005). HuR as a negative post-transcriptional modulator in inflammation. Mol. Cell 19, 777–789. doi: 10.1016/j.molcel.2005.08.007
Kedersha, N., Chen, S., Gilks, N., Li, W., Miller, I. J., Stahl, J., et al. (2002). Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195–210. doi: 10.1091/mbc.01-05-0221
Kostura, M., and Mathews, M. B. (1989). Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol. Cell. Biol. 9, 1576–1586. doi: 10.1128/MCB.9.4.1576
Leppek, K., Das, R., and Barna, M. (2017). Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158–174. doi: 10.1038/nrm.2017.103
Lu, P. D., Harding, H. P., and Ron, D. (2004). Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33. doi: 10.1083/jcb.200408003
Ma, X. M., and Blenis, J. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318. doi: 10.1038/nrm2672
Markmiller, S., Soltanieh, S., Server, K. L., Mak, R., Jin, W., Fang, M. Y., et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13. doi: 10.1016/j.cell.2017.12.032
Mayr, C. (2016). Evolution and biological roles of alternative 3′UTRs. Trends Cell Biol. 26, 227–237. doi: 10.1016/j.tcb.2015.10.012
Mayr, C. (2017). Regulation by 3′-untranslated regions. Annu. Rev. Genet. 51, 171–194. doi: 10.1146/annurev-genet-120116-024704
McCormick, C., and Khaperskyy, D. A. (2017). Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 17, 647–660. doi: 10.1038/nri.2017.63
Mizrahi, O., Nachshon, A., Shitrit, A., Gelbart, I. A., Dobesova, M., Brenner, S., et al. (2018). Virus-induced changes in mRNA secondary structure uncover cis-regulatory elements that directly control gene expression. Mol. Cell 72, 862–874.e5. doi: 10.1016/j.molcel.2018.09.003
Munn, D. H., Sharma, M. D., Baban, B., Harding, H. P., Zhang, Y., Ron, D., et al. (2005). GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22, 633–642. doi: 10.1016/j.immuni.2005.03.013
Namer, L. S., Osman, F., Banai, Y., Masquida, B., Jung, R., and Kaempfer, R. (2017). An ancient pseudoknot in TNF-α pre-mRNA activates PKR, inducing eIF2α phosphorylation that potently enhances splicing. Cell Rep. 20, 188–200. doi: 10.1016/j.celrep.2017.06.035
Naz, S., Battu, S., Khan, R. A., Afroz, S., Giddaluru, J., Vishwakarma, S. K., et al. (2019). Activation of integrated stress response pathway regulates IL-1β production through post-transcriptional and translational reprogramming in macrophages. Eur. J. Immunol. 49, 277–289. doi: 10.1002/eji.201847513
Osman, F., Jarrous, N., Ben-Asouli, Y., and Kaempfer, R. (1999). A cis-acting element in the 3'-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 13, 3280–3293. doi: 10.1101/gad.13.24.3280
Pakos-Zebrucka, K., Koryga, I., Mnich, K., Ljujic, M., Samali, A., and Gorman, A. M. (2016). The integrated stress response. EMBO Rep. 17, 1374–1395. doi: 10.15252/embr.201642195
Panas, M. D., Ivanov, P., and Anderson, P. (2016). Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215, 313–323. doi: 10.1083/jcb.201609081
Piecyk, M., Wax, S., Beck, A. R. P., Kedersha, N., Gupta, M., Maritim, B., et al. (2000). TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19, 4154–4163. doi: 10.1093/emboj/19.15.4154
Protter, D. S. W., and Parker, R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. doi: 10.1016/j.tcb.2016.05.004
Pulendran, B. (2015). The varieties of immunological experience: of pathogens, stress, and dendritic cells. Annu. Rev. Immunol. 33, 563–606. doi: 10.1146/annurev-immunol-020711-075049
Ravindran, R., Khan, N., Nakaya, H. I., Li, S., Loebbermann, J., Maddur, M. S., et al. (2014). Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 343, 313–317. doi: 10.1126/science.1246829
Ravindran, R., Loebbermann, J., Nakaya, H. I., Khan, N., Ma, H., Gama, L., et al. (2016). The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature 531, 523–527. doi: 10.1038/nature17186
Ribas, A., and Wolchok, J. D. (2018). Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355. doi: 10.1126/science.aar4060
Ricciardi, S., Manfrini, N., Alfieri, R., Calamita, P., Crosti, M. C., Gallo, S., et al. (2018). The translational machinery of human CD4+ T cells is poised for activation and controls the switch from quiescence to metabolic remodeling. Cell Metab. 28, 895–906.e5. doi: 10.1016/j.cmet.2018.08.009
Rodriguez, P. C., Hernandez, C. P., Morrow, K., Sierra, R., Zabaleta, J., Wyczechowska, D. D., et al. (2010). l-Arginine deprivation regulates cyclin D3 mRNA stability in human T cells by controlling HuR expression. J. Immunol. 185, 5198–5204. doi: 10.4049/jimmunol.1001224
Rodriguez, P. C., Quiceno, D. G., and Ochoa, A. C. (2007). l-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 109, 1568–1573. doi: 10.1182/blood-2006-06-031856
Saito, K., Chen, S., Piecyk, M., and Anderson, P. (2001). TIA-1 regulates the production of tumor necrosis factor alpha in macrophages, but not in lymphocytes. Arthritis Rheum. 44, 2879–2887. doi: 10.1002/1529-0131(200112)44:12<2879::AID-ART476>3.0.CO;2-4
Sandberg, R., Neilson, J. R., Sarma, A., Sharp, P. A., and Burge, C. B. (2008). Proliferating Cells Express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science 320, 1643–1647. doi: 10.1126/science.1155390
Scheu, S., Stetson, D. B., Reinhardt, R. L., Leber, J. H., Mohrs, M., and Locksley, R. M. (2006). Activation of the integrated stress response during T helper cell differentiation. Nat. Immunol 7, 644–651. doi: 10.1038/ni1338
Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., et al. (2001). Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176. doi: 10.1016/S1097-2765(01)00265-9
Schneider, K., Nelson, G. M., Watson, J. L., Morf, J., Dalglish, M., Luh, L. M., et al. (2020). Protein stability buffers the cost of translation attenuation following eIF2α phosphorylation. Cell Rep. 32:108154. doi: 10.1016/j.celrep.2020.108154
Schwanhäusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., et al. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. doi: 10.1038/nature10098
Sundrud, M. S., Koralov, S. B., Feuerer, M., Calado, D. P., Kozhaya, A. E., Rhule-Smith, A., et al. (2009). Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science 324, 1334–1338. doi: 10.1126/science.1172638
Tan, H., Yang, K., Li, Y., Shaw, T. I., Wang, Y., Blanco, D. B., et al. (2017a). Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503. doi: 10.1016/j.immuni.2017.02.010
Tan, T. C. J., Knight, J., Sbarrato, T., Dudek, K., Willis, A. E., and Zamoyska, R. (2017b). Suboptimal T-cell receptor signaling compromises protein translation, ribosome biogenesis, and proliferation of mouse CD8 T cells. Proc. Natl. Acad. Sci. U.S.A. 114, E6117–E6126. doi: 10.1073/pnas.1700939114
Taniuchi, S., Miyake, M., Tsugawa, K., Oyadomari, M., and Oyadomari, S. (2016). Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci. Rep. 6:32886. doi: 10.1038/srep32886
Van de Velde, L.-A., Guo, X.-Z. J., Barbaric, L., Smith, A. M., Oguin, T. H., Thomas, P. G., et al. (2016). Stress kinase GCN2 controls the proliferative fitness and trafficking of cytotoxic T cells independent of environmental amino acid sensing. Cell Rep. 17, 2247–2258. doi: 10.1016/j.celrep.2016.10.079
Vogel, K. U., Edelmann, S. L., Jeltsch, K. M., Bertossi, A., Heger, K., Heinz, G. A., et al. (2013). Roquin paralogs 1 and 2 redundantly repress the icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity 38, 655–668. doi: 10.1016/j.immuni.2012.12.004
Wolf, T., Jin, W., Zoppi, G., Vogel, I. A., Akhmedov, M., Bleck, C. K. E., et al. (2020). Dynamics in protein translation sustaining T cell preparedness. Nat. Immunol. 21, 927–937. doi: 10.1038/s41590-020-0714-5
Wong, W. L., Brostrom, M. A., Kuznetsov, G., Gmitter-Yellen, D., and Brostrom, C. O. (1993). Inhibition of protein synthesis and early protein processing by thapsigargin in cultured cells. Biochem. J. 289(Pt 1), 71–79. doi: 10.1042/bj2890071
Yang, X., Xia, R., Yue, C., Zhai, W., Du, W., Yang, Q., et al. (2018). ATF4 regulates CD4+ T cell immune responses through metabolic reprogramming. Cell Rep. 23, 1754–1766. doi: 10.1016/j.celrep.2018.04.032
Yu, C., York, B., Wang, S., Feng, Q., Xu, J., and O'Malley, B. W. (2007). An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol. Cell 25, 765–778. doi: 10.1016/j.molcel.2007.01.025
Yu, D., Tan, A. H.-M., Hu, X., Athanasopoulos, V., Simpson, N., Silva, D. G., et al. (2007). Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature 450, 299–303. doi: 10.1038/nature06253
Zhang, Q., and Vignali, D. A. A. (2016). Co-stimulatory and co-inhibitory pathways in autoimmunity. Immunity 44, 1034–1051. doi: 10.1016/j.immuni.2016.04.017
Keywords: stress granules (SG), immune cells, post-transcriptional regulation, translation, mRNA, RNA-binding proteins (RBP), integrated stress response (ISR), mTOR
Citation: Curdy N, Lanvin O, Cadot S, Laurent C, Fournié J-J and Franchini D-M (2021) Stress Granules in the Post-transcriptional Regulation of Immune Cells. Front. Cell Dev. Biol. 8:611185. doi: 10.3389/fcell.2020.611185
Received: 29 September 2020; Accepted: 07 December 2020;
Published: 14 January 2021.
Edited by:
Santosh Chauhan, Institute of Life Sciences (ILS), IndiaReviewed by:
Antje Ostareck-Lederer, University Hospital RWTH Aachen, GermanyJianbo Sun, Sun Yat-sen University, China
Copyright © 2021 Curdy, Lanvin, Cadot, Laurent, Fournié and Franchini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Don-Marc Franchini, don-marc.franchini@inserm.fr
 Nicolas Curdy
Nicolas Curdy Olivia Lanvin
Olivia Lanvin Sarah Cadot1,2,3
Sarah Cadot1,2,3  Camille Laurent
Camille Laurent Jean-Jacques Fournié
Jean-Jacques Fournié Don-Marc Franchini
Don-Marc Franchini