Phytochemical Constituents of Citrus hystrix DC. Leaves Attenuate Inflammation via NF-κB Signaling and NLRP3 Inflammasome Activity in Macrophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Plant Material Preparation and Extraction of CH Leaves
2.3. Isolation and Identification of Active Compounds
2.4. Primary Human Monocyte Isolation and Differentiation
2.5. Differentiation of THP-1 Cells and Inflammasome Activation
2.6. Flow Cytometry Analysis
2.7. Cell Viability Measurement
2.8. Evaluation of Anti-Inflammatory Effect
2.9. Enzyme-Linked Immunosorbent Assay (ELISA)
2.10. Real-Time Quantitative RT-PCR
2.11. SDS-PAGE and Western Blot Analysis
2.12. Statistical Analysis
3. Results
3.1. Extraction and Fractionation
3.2. Identification of an Active Compound Isolated from CHAF
3.3. Cell Surface Marker Analysis of Human MDMs by Flow Cytometry
3.4. CHAF and Lupeol Suppression on Production of Pro-Inflammatory Mediators in LPS-Stimulated Human MDMs
3.5. Effect of CHAF and Lupeol on NLRP3 Inflammasome mRNA Expression in THP-1
3.6. The Effect of CHAF and Lupeol on NF-κB Signaling and COX-2 Proteins
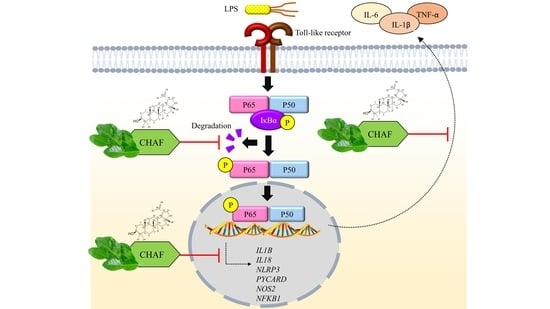
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newton, K.; Dixit, V.M. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2012, 4, a006049. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.S.; Sohn, D.H. Damage-Associated Molecular Patterns in Inflammatory Diseases. Immune Netw. 2018, 18, e27. [Google Scholar] [CrossRef]
- Arango Duque, G.; Descoteaux, A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front. Immunol. 2014, 5, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oishi, Y.; Manabe, I. Macrophages in inflammation, repair and regeneration. Int. Immunol. 2018, 30, 511–528. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.V.; Deng, M.; Ting, J.P.Y. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Maione, F.; Russo, R.; Khan, H.; Mascolo, N. Medicinal plants with anti-inflammatory activities. Nat. Prod. Res. 2016, 30, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Hutadilok-Towatana, N.; Chaiyamutti, P.; Panthong, K.; Mahabusarakam, W.; Rukachaisirikul, V. Antioxidative and Free Radical Scavenging Activities of Some Plants Used in Thai Folk Medicine. Pharm. Biol. 2006, 44, 221–228. [Google Scholar] [CrossRef]
- Agouillal, F.; Taher, Z.; Moghrani, H.; Nasrallah, N.; El Enshasy, H. A Review of Genetic Taxonomy, Biomolecules Chemistry and Bioactivities of Citrus hystrix DC. Biosci. Biotechnol. Res. Asia 2017, 14, 285–305. [Google Scholar] [CrossRef]
- Putri, H.; Nagadi, S.; Larasati, Y.A.; Wulandari, N.; Hermawan, A.; Nugroho, A.E. Cardioprotective and hepatoprotective effects of Citrus hystrix peels extract on rats model. Asian Pac. J. Trop. Biomed. 2013, 3, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Kooltheat, N.; Kamuthachad, L.; Anthapanya, M.; Samakchan, N.; Sranujit, R.P.; Potup, P.; Ferrante, A.; Usuwanthim, K. Kaffir lime leaves extract inhibits biofilm formation by Streptococcus mutans. Nutrition 2016, 32, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Pumival, P.; Tadtong, S.; Athikomkulchai, S.; Chittasupho, C. Antifungal Activity and the Chemical and Physical Stability of Microemulsions Containing Citrus hystrix DC Leaf Oil. Nat. Prod. Commun. 2020, 15, 1934578X20957755. [Google Scholar] [CrossRef]
- Kidarn, S.; Saenjum, C.; Hongwiset, D.; Phrutivorapongkul, A. Furanocoumarins from Kaffir lime and their inhibitory effects on inflammatory mediator production. Cogent Chem. 2018, 4, 1529259. [Google Scholar] [CrossRef]
- Kim, J.; Ahn, H.; Han, B.-C.; Shin, H.; Kim, J.-C.; Jung, E.-M.; Kim, J.; Yang, H.; Lee, J.; Kang, S.G.; et al. Obovatol inhibits NLRP3, AIM2, and non-canonical inflammasome activation. Phytomedicine 2019, 63, 153019. [Google Scholar] [CrossRef] [PubMed]
- Präbst, K.; Engelhardt, H.; Ringgeler, S.; Hübner, H. Basic Colorimetric Proliferation Assays: MTT, WST, and Resazurin. In Cell Viability Assays: Methods and Protocols; Gilbert, D.F., Friedrich, O., Eds.; Springer: New York, NY, USA, 2017; pp. 1–17. [Google Scholar]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013, 3, 71–85. [Google Scholar] [PubMed]
- Jain, P.; Bari, S. Isolation of Lupeol, Stigmasterol and Campesterol from Petroleum Ether Extract of Woody Stem of Wrightia tinctoria. Asian J. Plant Sci. 2010, 9, 163–167. [Google Scholar] [CrossRef] [Green Version]
- Shwe, H.H.; Win, K.K.; Moe, T.T.; Myint, A.A.; Win, T. Isolation and Structural Characterization of Lupeol from the Stem Bark of Diospyros ehretioides Wall. IEEE-SEM 2019, 7, 140–144. [Google Scholar]
- Ragasa, C.; Tan, M.; Fortin, D.; Shen, C.-C. Chemical Constituents of Ixora philippinensis Merr. J. Appl. Pharm. Sci. 2015, 5, 62–67. [Google Scholar] [CrossRef] [Green Version]
- Lertsatitthanakorn, P.; Taweechaisupapong, S.; Aromdee, C.; Khunkitti, W. In vitro bioactivities of essential oils used for acne control. Int. J. Aromather. 2006, 16, 43–49. [Google Scholar] [CrossRef]
- Murakami, A.; Gao, G.; Kim, O.K.; Omura, M.; Yano, M.; Ito, C.; Furukawa, H.; Jiwajinda, S.; Koshimizu, K.; Ohigashi, H. Identification of Coumarins from the Fruit of Citrus hystrix DC as Inhibitors of Nitric Oxide Generation in Mouse Macrophage RAW 264.7 Cells. J. Agric. Food Chem. 1999, 47, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Sakai, J.; Cammarota, E.; Wright, J.A.; Cicuta, P.; Gottschalk, R.A.; Li, N.; Fraser, I.D.C.; Bryant, C.E. Lipopolysaccharide-induced NF-κB nuclear translocation is primarily dependent on MyD88, but TNFα expression requires TRIF and MyD88. Sci. Rep. 2017, 7, 1428. [Google Scholar] [CrossRef]
- Xu, M.; Li, X.; Song, L.; Tao, C.; Fang, J.; Tao, L. Lupeol alleviates coxsackievirus B3-induced viral myocarditis in mice via downregulating toll-like receptor 4. J. Int. Med Res. 2020, 48, 030006052091090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira-Junior, M.S.; Pereira, E.P.; de Amorim, V.C.M.; Reis, L.T.C.; do Nascimento, R.P.; da Silva, V.D.A.; Costa, S.L. Lupeol inhibits LPS-induced neuroinflammation in cerebellar cultures and induces neuroprotection associated to the modulation of astrocyte response and expression of neurotrophic and inflammatory factors. Int. Immunopharmacol. 2019, 70, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Weigenand, O.; Hussein, A.A.; Lall, N.; Meyer, J.J.M. Antibacterial Activity of Naphthoquinones and Triterpenoids from Euclea natalensis Root Bark. J. Nat. Prod. 2004, 67, 1936–1938. [Google Scholar] [CrossRef] [PubMed]
- Ajaiyeoba, E.O.; Ashidi, J.S.; Okpako, L.C.; Houghton, P.J.; Wright, C.W. Antiplasmodial compounds from Cassia siamea stem bark extract. Phytother. Res. 2008, 22, 254–255. [Google Scholar] [CrossRef] [PubMed]
- Pitchai, D.; Roy, A.; Ignatius, C. In vitro evaluation of anticancer potentials of lupeol isolated from Elephantopus scaber L. on MCF-7 cell line. J. Adv. Pharm. Technol. Res. 2014, 5, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Okusa, P.N.; Stévigny, C.; Névraumont, M.; Gelbcke, M.; Van Antwerpen, P.; Braekman, J.C.; Duez, P. Ferulaldehyde and lupeol as direct and indirect antimicrobial compounds from Cordia gilletii (Boraginaceae) root barks. Nat. Prod. Commun. 2014, 9, 619–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wal, P.; Awani, R.; Wal, A.; Sharma, G. Biological activities of lupeol. Syst. Rev. Pharm. 2011, 2, 96. [Google Scholar] [CrossRef]
- Théophile, D.; Laure, N.E.; Benoît, N.T.; Anatole, A.G.B.; Emmanuel, A.A.; Paul, T.V.; Pierre, K. Antinociceptive and anti-inflammatory effects of the ethyl acetate stem bark extract of Bridelia scleroneura (Euphorbiaceae). Inflammopharmacology 2006, 14, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Ongoka, P.; Banzouzi, J.; Poupat, C.; Ouamba, J. Steroids isolated from Millettia versicolor Baker (Fabaceae). Afr. J. Biotechnol. 2008, 7, 1727–1730. [Google Scholar] [CrossRef] [Green Version]
- Anuchapreeda, S.; Chueahongthong, F.; Viriyaadhammaa, N.; Panyajai, P.; Anzawa, R.; Tima, S.; Ampasavate, C.; Saiai, A.; Rungrojsakul, M.; Usuki, T.; et al. Antileukemic Cell Proliferation of Active Compounds from Kaffir Lime (Citrus hystrix) Leaves. Molecules 2020, 25, 1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geetha, T.; Varalakshmi, P. Anti-inflammatory activity of lupeol and lupeol linoleate in rats. J. Ethnopharmacol. 2001, 76, 77–80. [Google Scholar] [CrossRef]
- Bani, S.; Kaul, A.; Khan, B.; Fayaz Ahmad, S.; Suri, K.A.; Gupta, B.; Satti, N.; Qazi, G. Suppression of T lymphocyte activity by lupeol isolated from Crataeva religiosa. Phytother. Res. PTR 2006, 20, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, J.F.; Teixeira, M.M.; Barbosa-Filho, J.M.; Lúcio, A.S.S.C.; Almeida, J.R.G.S.; de Queiroz, L.P.; Ribeiro-dos-Santos, R.; Soares, M.B.P. The triterpenoid lupeol attenuates allergic airway inflammation in a murine model. Int. Immunopharmacol. 2008, 8, 1216–1221. [Google Scholar] [CrossRef] [PubMed]







| Gene | Description | Forward (5′ → 3′) | Reverse (5′ → 3′) |
|---|---|---|---|
| IL1B | Interleukin 1 beta | AGCTACGAATCTCCGACCAC | CGTTATCCCATGTGTCGAAGAA |
| IL18 | Interleukin 18 | GAAGATGCCAGGGGTAATGA | TACCTGCCCCAAACTGAAAC |
| CASP1 | Caspase-1 | CTTGCTTGAAATGTGCTCCA | AGTGGCATCCCTGTTTGTTC |
| NLRP3 | NLR family pyrin domain containing 3 | ACAAACTCATGGTGGCTTCC | GGCCAGAAGAAAAGCAAGTG |
| PYCARD | PYD and CARD domain containing | TGACGGATGAGCAGTACCAG | AGGATGATTTGGTGGGATTG |
| NFKB1 | Nuclear factor kappa B subunit 1 | AACAGAGAGGATTTCGTTT | TTTGACCTGAGGGTAAGAC |
| NOS2 | Nitric oxide synthase 2 | TTCAGTATCACAACCTCAGCAAG | TGGACCTGCAAGTTAAAAT |
| ACTB | Actin beta | AGAAAATCTGGCACCACACC | CCATCTCTTGCTCGAAGTCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buakaew, W.; Pankla Sranujit, R.; Noysang, C.; Thongsri, Y.; Potup, P.; Nuengchamnong, N.; Suphrom, N.; Usuwanthim, K. Phytochemical Constituents of Citrus hystrix DC. Leaves Attenuate Inflammation via NF-κB Signaling and NLRP3 Inflammasome Activity in Macrophages. Biomolecules 2021, 11, 105. https://doi.org/10.3390/biom11010105
Buakaew W, Pankla Sranujit R, Noysang C, Thongsri Y, Potup P, Nuengchamnong N, Suphrom N, Usuwanthim K. Phytochemical Constituents of Citrus hystrix DC. Leaves Attenuate Inflammation via NF-κB Signaling and NLRP3 Inflammasome Activity in Macrophages. Biomolecules. 2021; 11(1):105. https://doi.org/10.3390/biom11010105
Chicago/Turabian StyleBuakaew, Watunyoo, Rungnapa Pankla Sranujit, Chanai Noysang, Yordhathai Thongsri, Pachuen Potup, Nitra Nuengchamnong, Nungruthai Suphrom, and Kanchana Usuwanthim. 2021. "Phytochemical Constituents of Citrus hystrix DC. Leaves Attenuate Inflammation via NF-κB Signaling and NLRP3 Inflammasome Activity in Macrophages" Biomolecules 11, no. 1: 105. https://doi.org/10.3390/biom11010105