Abstract
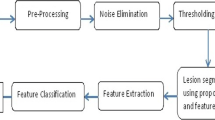
Lesion segmentation is a vital step in a melanoma recognition system. Many algorithms were developed for the efficient skin lesion segmentation. Most of them fails to realize a perfect segmentation. This paper proposes a novel, fully automatic system, for the lesion segmentation in dermatograms. The proposed approach executes in two steps. Selection of root seed is the first step. All the lesion pixels in the dermatogram are identified during the second step. Traversal through a predefined lesion pixel path ensures the reachability of all lesion pixels irrespective of the possible lesion discontinuity. The proposed algorithm is tested with two publically available dataset, PH2 and images of ISBI2016 challenge. Out of the six evaluation parameters, the proposed method shows the best values for specificity, accuracy, Hammuode distance and XOR. This confirms the merit of the proposal with respect to existing popular methods.






Similar content being viewed by others
REFERENCES
Cancer.org. Key Statistics for Melanoma Skin Cancer (2018). https://www.cancer.org/cancer/melanoma-skin-cancer/about/key-statistics.html. Accessed September 14, 2018.
O. Abuzaghleh, B. D. Barkana, and M. Faezipour, “Noninvasive real-time automated skin lesion analysis system for melanoma early detection and prevention,” IEEE J. Trans. Eng. Health Med. 3, 2900310 (2015). https://doi.org/10.1109/JTEHM.2015.2419612
J. Glaister, A. Wong, and D. Clausi, “Segmentation of skin lesions from digital images using joint statistical texture distinctiveness,” IEEE Trans. Biomed. Eng. 61 (4), 1220–1230 (2014).
M. Silveira et al., “Comparison of segmentation methods for melanoma diagnosis in dermoscopy images,” IEEE J. Sel. Top. Signal Process. 3, 35–45 (2009).
M. E. Celebi, G. Schaefer, H. Iyatomi, and W. V. Stoecker., “Lesion border detection in dermoscopy images,” Comput. Med. Imaging Graphics 33, 148–153 (2009).
O. B. Akinrinade, P. Adewale Owolawi, C. Tu, and T. Mapayi, “Graph-cuts technique for melanoma segmentation over different color spaces,” in 2018 International Conference on Intelligent and Innovative Computing Applications (ICONIC) (2018), pp. 1–5. https://doi.org/10.1109/ICONIC.2018.8601269
F. Melgani, “Robust image binarization with ensembles of thresholding algorithms,” J. Electron. Imaging 15 (2), 023010 (2008). https://doi.org/10.1117/1.2194767
M. Emre Celebi et al., “Border detection in dermoscopy images using statistical region merging,” Skin Res. Technol. 14, 347–353 (2008).
J. Wang, H. Jiang, Z. Yuan, M.-M. Cheng, X. Hu, and N. Zheng, “Salient object detection: A discriminative regional feature integration approach,” Int. J. Comput. Vis. 123 (2), 251–268 (2017).
M. Jahanifar, N. Zamani Tajeddin, B. Mohammadzadeh Asl, and A. Gooya, “Supervised saliency map driven segmentation of lesions in dermoscopic images,” IEEE J. Biomed. Health Inf. 23 (2), 509–518 (2019). https://doi.org/10.1109/JBHI.2018.2839647
M. Attia, M. Hossny, S. Nahavandi, and A. Yazdabadi, “Skin melanoma segmentation using recurrent and convolutional neural networks,” in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) (Melbourne, 2017), pp. 292–296. https://doi.org/10.1109/ISBI.2017.7950522
L. Yu, H. Chen, Q. Dou, J. Qin, and P. Heng, “Automated melanoma recognition in dermoscopy images via very deep residual networks,” IEEE Trans. Med. Imaging 36 (4), 994–1004 (2017). https://doi.org/10.1109/TMI.2016.2642839
L. Bi, J. Kim, E. Ahn, D. Feng, and M. Fulham, “Automatic melanoma detection via multi-scale lesion-biased representation and joint reverse classification,” in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) (Prague, 2016), pp. 1055–1058. https://doi.org/10.1109/ISBI.2016.7493447
S. Pathan, K. Prabhu, and P. Siddalingaswamy, “Hair detection and lesion segmentation in dermoscopic images using domain knowledge,” Med. Biol. Eng. Comput. 56 (11), 2051–2065 (2018). https://doi.org/10.1007/s11517-018-1837-9
H. Fan, F. Xie, Y. Li, et al., “Automatic segmentation of dermoscopy images using saliency combined with Otsu threshold,” Comput. Biol. Med. 85, 75–85 (2017). https://doi.org/10.1016/j.compbiomed.2017.03.025
F. Riaz, H. Farhan, A. Hassan, M. Javed, and M. Coimbra, “Detecting melanoma in dermoscopy images using scale adaptive local binary patterns,” in 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC 2014) (2014), pp. 6758–6761. https://doi.org/10.1109/EMBC.2014.6945179
M. Khan et al., “An implementation of normal distribution based segmentation and entropy controlled features selection for skin lesion detection and classification,” BMC Cancer 18 (1), (2018). https://doi.org/10.1186/s12885-018-4465-8
O. B. Akinrinade, P. Adewale Owolawi, C. Tu, and T. Mapayi, “Graph-cuts technique for melanoma segmentation over different color spaces,” in 2018 International Conference on Intelligent and Innovative Computing Applications (ICONIC) (Plaine Magnien, 2018), pp. 1–5. https://doi.org/10.1109/ICONIC.2018.8601269
F. Riaz, S. Naeem, R. Nawaz, and M. Coimbra, “Active contours based segmentation and lesion periphery analysis for characterization of skin lesions in dermoscopy images,” IEEE J. Biomed. Health Inf. 23 (2), 489–500 (2019). https://doi.org/10.1109/JBHI.2018.2832455
T. Mendonca, P. M. Ferreira, J. S. Marques, A. R. Marcal, and J. Rozeira, “Ph2—a dermoscopic image database for research and benchmarking,” Int. Conf. IEEE Eng. Med. Biol. Soc. 2013, 5437–5440 (2013). http://www.fc.up.pt/addi/ph2.
D. Gutman et al., “Skin lesion analysis toward melanoma detection: A challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC)” (2016). arXiv:1605.01397.
W. R. Crum et al., “Generalized overlap measures for evaluation and validation in medical image analysis,” IEEE Trans. Med. Imaging 25, 1451–1461 (2006).
A. Hammoude, “Computer-assisted endocardial border identification from a sequence of two-dimensional echocardiographic images,” Thesis (Comput. Sci. Dept., Univ. Wash., 1988).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest. The authors declare that they have no conflict of interest.
Ethical approval. This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information

Nikesh P. received his B Tech in Computer Science and Engineering (2009) from the Kannur university in kerala and M Tech in Computer Cognition Technology (2012) from University of Mysore. He is currently an assistant professor in the Computer Science and Engineering Department at Government Engineering College Wayanad. His research interests include artificial intelligence, image processing and video processing.

Dr. Raju G. is currently working as Professor in the Department of Data Science, Christ (Deemed to be University), Lavasa, Pune, India. He obtained his masters as well as doctoral degrees from University of Kerala, India. His area of research includes Image Processing, Computer Vision, Machine Learning and Data Analytics. Prior to joining Christ, he worked in Kannur University, Kerala, Government Colleges, Kerala and Waljat Colleges, Sultanate of Oman. He successfully guided eighteen Ph. D. students and also published more than hundred research papers.
Rights and permissions
About this article
Cite this article
Nikesh, P., Raju, G. Automatic Skin Lesion Segmentation—A Novel Approach of Lesion Filling through Pixel Path. Pattern Recognit. Image Anal. 30, 815–826 (2020). https://doi.org/10.1134/S1054661820040215
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1054661820040215