Abstract
Background
Avr-Pita was the first effector identified in the blast fungus (Magnaporthe oryzae)–rice (Oryza sativa) pathosystem. However, the molecular mechanism underlying its effects on the host plant has remained a long-standing mystery.
Results
Here, we report that ectopically expressing Avr-Pita in rice enhances susceptibility to M. oryzae and suppresses pathogen-associated molecular pattern (PAMP)-triggered defense responses. Avr-Pita targets the host mitochondria and interacts with the cytochrome c oxidase (COX) assembly protein OsCOX11, a key regulator of mitochondrial reactive oxygen species (ROS) metabolism in rice. Overexpressing Avr-Pita or OsCOX11 increased COX activity and decreased ROS accumulation triggered by the fungal PAMP chitin. OsCOX11-overexpressing plants showed increased susceptibility to M. oryzae, whereas OsCOX11-knockdown plants showed resistance to M. oryzae.
Conclusions
Taken together, these findings suggest that the fungal pathogen M. oryzae delivers the effector Avr-Pita to the host plant, where it enhances COX activity thus decreasing ROS accumulation. Therefore, this effector suppresses host innate immunity by perturbing ROS metabolism in the mitochondria.
Similar content being viewed by others
Introduction
Plants have evolved sophisticated mechanisms to perceive and respond to pathogen attack, such as pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Liu et al., 2014). PTI and ETI are associated with immune responses including the rapid influx of calcium, a burst of reactive oxygen species (ROS), mitogen activated protein kinase (MAPK) phosphorylation cascades, callose deposition, pathogenesis-related (PR) gene expression, and the biosynthesis of antimicrobial compounds (Bigeard et al., 2015).
Rice blast caused by the hemibiotrophic fungal pathogen Magnaporthe oryzae (M. oryzae) is a devastating disease that threatens global food security (Dean et al., 2012; Skamnioti & Gurr, 2009). The rice–M. oryzae pathosystem is a well-known model for studying plant–pathogen interactions (Wilson & Talbot, 2009). Over the past decades, several M. oryzae avirulence (Avr) effector genes have been cloned and characterized, including PWL1, PWL2, PWL3, PWL4, Avr-Pita, ACE1, AvrPiz-t, Avr-Pia, Avr-Pii, Avr-Pik/km/kp, Avr1-CO39, Avr-Pi9, and Avr-Pib (Zhang & Xu, 2014; Zhang et al., 2018). However, the virulence functions of only a few Avr proteins have been elucidated. AvrPiz-t manipulates host immunity by interacting with the RING-type ubiquitin E3 ligases APIP6 and APIP10, thereby inhibiting their E3 ligase activity and promoting their degradation, whereas APIP6 and APIP10 degrade AvrPiz-t (Park et al., 2012; Park et al., 2016). Inside the host cell, AvrPiz-t targets the bZIP-type transcription factor APIP5 to eliminate APIP5-triggered cell death and promote tissue necrosis (Wang et al., 2016). AvrPiz-t also interacts with the potassium channel OsAKT1 by competing with the cytosolic protein kinase OsCIPK23 to modulate K+ channel activity (Shi et al., 2018). Moreover, AvrPiz-t interacts with the nucleoporin protein APIP12 to suppress host immunity (Tang et al., 2017). Another well-documented Avr protein, AvrPii, disrupts host immune systems by interacting with NADP-MALIC ENZYME 2 (OsNADP-ME2) to inhibit NADP-ME activity or by interacting with two exocytosis complex OsExo70 proteins (Fujisaki et al., 2015; Singh et al., 2016). These evidences indicate that Avr proteins may target to various host factors to manipulate host immunity. However, the function of Avr protein is still largely unknown.
Although Avr-Pita was the first fungal effector isolated from the M. oryzae–rice pathosystem (Jia et al., 2000; Orbach et al., 2000), our mechanistic understanding of its role in this pathosystem remains limited. Avr-Pita is a putative neutral zinc metalloprotease with a signal peptide at its N-terminus that is thought to be recognized by the nucleotide binding site-leucine rich repeat domain (NBS-LRR) of protein Pi-ta, which triggers host immune responses in rice cells (Bryan et al., 2000; Orbach et al., 2000). During the infection processes, Avr-Pita accumulates in the biotrophic interfacial complex (BIC) structure and is delivered into host cells via invasive M. oryzae hyphae (Khang et al., 2010). However, where and how Avr-Pita functions inside host cells remains unclear.
ROS play key roles in plant defense signaling. In response to the perception of pathogen invasion, plants activate a rapid ROS burst leading to the hypersensitive response (HR), programmed cell death (PCD), and kinase cascade (Smirnoff & Arnaud, 2019). High concentrations of ROS also function as toxicants to the pathogen (Kou et al., 2019). ROS are produced in several cellular compartments including the plasma membrane, mitochondria, peroxisomes, and chloroplasts (Qi et al., 2018; Smirnoff & Arnaud, 2019). The generation of ROS by the plasma membrane enzyme NADPH oxidase during plant defense responses has been well documented. The mitochondrion is a major organelle that produces ROS via Complexes I, II, and III of the mitochondrial electron transport chain (METC) (Vanlerberghe, 2013). However, little is known about the association of mitochondrial METC Complexes I and III with defense responses (Vidal et al., 2007).
Here, we demonstrate that Avr-Pita targets host mitochondria and interacts with cytochrome c oxidase (COX) assembly protein OsCOX11 of the METC, which promotes the activity of Complex IV of the METC, thereby inhibiting ROS accumulation and suppressing innate immunity in rice.
Results
The Avr-Pita Effector Suppresses Defense Responses in Rice
To investigate the function of Avr-Pita inside rice cells, we expressed Avr-Pita in rice cells. To this end, we generated rice suspension cultures harboring the estradiol-induced construct PXVE::Avr-Pita (Fig. S1a). Estradiol treatment significantly induced the expression of Avr-Pita in these lines (Fig. S1c and S1e). The expression of the early defense response gene PHENYLALANINE AMMONIA LYASE 1 (OsPAL1) was markedly suppressed in response to estradiol treatment in these lines at the indicated time points (Fig. S1f), suggesting that inducing the expression of Avr-Pita inhibits basic defense responses in rice cells.
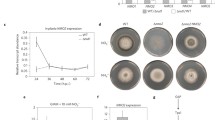
We also generated stably transformed PUbi::Avr-Pita rice plants overexpressing Avr-Pita under the control of the maize (Zea mays) Ubiquitin promoter (Fig. S1a). Although the overexpression of Avr-Pita in the leaves of PUbi::Avr-Pita plants was confirmed by quantitative reverse-transcription PCR (qRT-PCR) (Fig. S1d and S1g), ectopic overexpression of Avr-Pita did not significantly affect the morphology of rice plants in terms of growth and development (Fig. S1b). To test pathogen resistance, we inoculated the PUbi::Avr-Pita lines with M. oryzae isolate 10–441 (without Avr-Pita) by punch inoculation. The PUbi::Avr-Pita plants had larger disease lesions and higher relative fungal biomass than wild-type (WT) plants at 12 days post inoculation (dpi) (Fig. 1a–c). These results indicate that ectopic expression of Avr-Pita enhances rice susceptibility to M. oryzae.
Overexpressing Avr-Pita suppresses basal resistance to M. oryzae in rice. a Disease symptoms of PUbi::Avr-Pita lines and wild-type (WT) plants at 12 days post inoculation (dpi) with compatible isolate 10–441. b Lesion area was photographed and measured using ImageJ software. Data are shown as mean ± standard error (SD) (**P < 0.01, n > 12). c Relative fungal biomass of M. oryzae in inoculated leaves, as quantified by qPCR comparing the DNA amounts of the fungal gene MoPot2 and the rice gene OsUbq. Data are shown as mean ± SD, (**P < 0.01, n = 3). d Expression of OsPBZ1 in the leaves of PUbi::Avr-Pita lines following chitin treatment. e Expression of OsPR10 in PUbi::Avr-Pita lines following chitin treatment. Data were normalized to the expression level of OsACTIN. Data are shown as mean ± SD (**P < 0.01, n = 3)
To further investigate the PAMP-triggered defense response in rice, we treated the PUbi::Avr-Pita lines with chitin and monitored the expression of PR genes. The expression of the PR genes PROBENAZOLE-INDUCIBLE 1 (OsPBZ1) and OsPR10 was significantly suppressed in PUbi::Avr-Pita vs. WT plants, as indicated by qRT-PCR (Fig. 1d and e), indicating that Avr-Pita suppresses chitin-triggered PTI in rice.
Avr-Pita Interacts with OsCOX11 in Rice Mitochondria
To explore how Avr-Pita suppresses defense responses in plants, we conducted yeast two-hybrid (Y2H) assays using Avr-Pita49–224 (processed mature protease) as bait and identified six candidate proteins (Table S1). We selected the mitochondrial COX assembly protein, OsCOX11, for further study. The interaction between Avr-Pita and OsCOX11 was further validated using full-length Avr-Pita protein in the Y2H system (Fig. 2a). We confirmed the interaction of OsCOX11 and Avr-Pita via a pull-down assay using His:OsCOX11 and GST:Avr-Pita (Fig. 2b).
Avr-Pita interacts with OsCOX11. a Analysis of protein–protein interactions between BD:Avr-Pita (mature protease) and AD:OsCOX11 via yeast-two-hybrid (Y2H) assay. Yeast cells harboring the indicated bait and prey plasmids were diluted 10×, 100×, or 1000× and spotted onto selective media SD-LW and SD-AHLW. b In vitro pull-down assays were conducted with fusion proteins (GST:Avr-Pita and His:OsCOX11) using His-Ni beads and detected by immunoblot analysis with anti-GST and anti-His antibodies. Asterisks indicate the target protein. (c) Deletion assays of OsCOX11 to test interactions with Avr-Pita. A series of truncated OsCOX11 proteins were tested for interaction with Avr-Pita via Y2H
To narrow down the specific region of OsCOX11 that physically interacts with Avr-Pita, we generated a series of truncated OsCOX11 fragments for Y2H assays. The OsCOX11140–220 fragment was identified as a sufficient region responsible for the interaction with Avr-Pita, while OsCOX11199–220 fragment (containing the 5th β sheet region) was a critical region responsible for the interaction (Fig. 2c). However, no orthologs of OsCOX11, such as AtCOX11 (Arabidopsis thaliana, A. thaliana), ScCOX11 (Saccharomyces cerevisiae, S. cerevisiae), and MoCOX11 (M. oryzae), were able to interact with Avr-Pita (Fig. S2a and S2b). These results suggest that fungal Avr-Pita specifically binds to rice OsCOX11 via the specific region of OsCOX11.
To explore the subcellular localization of Avr-Pita inside plant cells, we generated rice protoplasts and onion epidermal cells transiently expressing Avr-Pita:yellow fluorescent protein (YFP) and examined them by confocal microscopy. Avr-Pita:YFP perfectly co-localized with the mitochondrial marker Rflb:mCherry (Wang et al., 2006) in rice protoplasts and with the mitochondrial dye MitoTracker in onion epidermal cells (Fig. 3a and S3a). Avr-Pita:YFP also co-localized with OsCOX11:mCherry in mitochondria (Fig. 3b and S3b). These results indicate that the fungal effector Avr-Pita targets the host mitochondria and binds OsCOX11, a subunit of the COX complex.
Avr-Pita and OsCOX11 co-localize to rice mitochondria. a Avr-Pita:YFP and mitochondrial marker Rf1b:mCherry were co-expressed in rice protoplasts. YFP and mCherry fluorescent signals overlapped in the mitochondria. b Avr-Pita:YFP and OsCOX11:mCherry were co-expressed in rice protoplasts. YFP and mCherry fluorescent signals overlapped in the mitochondria. Scale bars = 10 μm
OsCOX11 Negatively Regulates Disease Resistance
To investigate the role of OsCOX11 in plant resistance to M. oryzae, we generated OsCOX11-overexpressing transgenic lines (harboring the PUbi::OsCOX11 construct) for functional studies (Fig. S4a). When the PUbi::OsCOX11 plants were inoculated with M. oryzae strain 10–441, they developed larger disease lesions (Fig. 4a and b) and more fungal biomass (Fig. 4c) than the control at 12 dpi.
OsCOX11 functions as a negative regulator of basal resistance against M. oryzae. a Disease symptoms of PUbi::OsCOX11 and WT plants in response to M. oryzae isolate 10–441. The inoculated leaves were photographed at 12 dpi. b Lesion area was measured with ImageJ software. Data are shown as the mean ± SD (**P < 0.01, n > 12). c Fungal biomass in inoculated leaves, as quantified by qPCR comparing the DNA amounts of the fungal gene MoPot2 and the rice gene OsUbq. Data are shown as mean ± SD (**P < 0.01, n > 12). d Disease symptoms of oscox11/OsCOX11 and WT plants inoculated with M. oryzae isolate 13–219. The leaves were photographed at 12 dpi. e Lesion area was measured with ImageJ software. Data are shown as mean ± standard error (SD) (**P < 0.01, n > 12). f Relative fungal biomass on M. oryzae-inoculated leaves, as measured by qPCR. Data are shown as mean ± SD (**P < 0.01, n > 12)
To determine whether OsCOX11 plays a role in disease resistance, we produced OsCOX11-silenced and -knockout plants by RNAi and CRISPR/Cas9-mediated gene editing, respectively. Surprisingly, we failed to obtain homozygous knockout mutants (data not shown), indicating that OsCOX11 plays a crucial role in plant growth and development and that the homozygous mutant may be lethal. Therefore, we used the heterozygous OsCOX11 knockout mutant (oscox11/OsCOX11) for functional analysis (Fig. S4b and S4c) of disease resistance. Following inoculation, the disease lesions of oscox11/OsCOX11 plants were smaller than those of WT plants (Fig. 4d and e), indicating that oscox11/OsCOX11 plants are resistant to M. oryzae. Consistent with this, the OsCOX11-RNAi lines were also more resistant to M. oryzae than WT plants (Fig. S4d, S4e and S4f). Moreover, the fungal biomass was significantly reduced in the oscox11/OsCOX11 and OsCOX11-RNAi lines (Fig. 4f and S4g), indicating that OsCOX11 is a negative regulator of plant resistance to M. oryzae.
Avr-Pita Hampers Plant Resistance by Promoting the COX Activity in ROS Metabolism
To understand the effect of the relationship between Avr-Pita and OsCOX11 on plant resistance, we examined chitin-induced ROS accumulation in various transgenic plants using a luminol-based chemiluminescence assay. ROS accumulation was significantly suppressed in PUbi::Avr-Pita and PUbi::OsCOX11 plants, whereas the ROS burst was strongly delayed in oscox11/OsCOX11 cells after chitin treatment (Fig. 5a). Staining with 3, 3′-diaminobenzidine (DAB) also showed that pathogen-triggered ROS accumulation was markedly reduced in PUbi::Avr-Pita and Pubi::OsCOX11 vs. WT plants (Fig. 5b). These results indicate that Avr-Pita and OsCOX11 function in pathogen infection by suppressing ROS accumulation.
Overexpressing Avr-Pita or OsCOX11 suppresses chitin-triggered ROS accumulation in rice. a Chitin-induced ROS accumulation in WT, PUbi::Avr-Pita, PUbi::OsCOX11, and oscox11/OsCOX11 cells, as measured using a luminol-chemiluminescence assay. Data are shown as mean ± SD (**P < 0.01, n = 4). b ROS accumulation detected with DAB staining in WT, PUbi::Avr-Pita, PUbi::OsCOX11, and oscox11/OsCOX11 plants after inoculation with M. oryzae. The images were taken at 0, 24, and 48 h post inoculation (hpi). Scale bars = 50 μm
To investigate whether Avr-Pita manipulates OsCOX11-mediated ROS metabolism at the transcriptional level, we measured the expression of OsCOX11 in plants after chitin treatment and fungal infection. The expression of OsCOX11 did not significantly change after chitin treatment in PUbi::Avr-Pita or WT plants (Fig. S5a). And the expression of OsCOX11 was induced slightly only at 6 h post M. oryzae infection (Fig. S5b). These results suggest that Avr-Pita does not significantly promote OsCOX11 at the transcriptional level.
Given that COX assembly proteins are present in METC Complex IV, we examined whether the interaction of Avr-Pita with COX11 affects COX activity. COX activity was much higher in PUbi::Avr-Pita and PUbi::OsCOX11 plants than in the WT both before and after chitin treatment (Fig. 6). These results suggest that the effect of Avr-Pita overexpression is equivalent to that of OsCOX11, as overexpressing either gene promoted COX activity to reduce ROS accumulation.
Based on these results, we propose a working model for the role of the effector Avr-Pita in disturbing host innate immunity. According to our model, M. oryzae secretes Avr-Pita into rice cells through the BIC structure of invasive hyphae during rice cell invasion. Avr-Pita targets the host mitochondria and interacts with the COX assembly protein OsCOX11 to promote COX activity, leading to reduced ROS accumulation and increased susceptibility to the pathogen (Fig. 7).
A working model for the role of Avr-Pita in regulating defense responses in rice. The PRR on the rice plasma membrane perceives PAMP molecules from the fungal pathogen (M. oryzae) and activates PTI resulting in a ROS burst and HR to stop microbial invasion (left). In M. oryzae isolates carrying the effector Avr-Pita (right), this effector is secreted from M. oryzae and delivered into rice cells. There, Avr-Pita targets the host mitochondria and binds to OsCOX11 to promote COX activity, thereby decreasing ROS accumulation, resulting in reduced resistance to M. oryzae
Discussion
During plant–pathogen interactions, the pathogens deliver a number of effectors into the host cells to disturb host innate immunity and facilitate their own propagation (Zhang & Xu, 2014). In-depth studies of bacteria–plant interactions have revealed that bacterial pathogens manipulate host immunity via their effectors (Deslandes & Rivas, 2012). By contrast, little is known about the roles of fungal effectors in this process. For example, the localization and intrinsic function of Avr-Pita in host cells have remained long-standing questions following the discovery of the Avr-Pita/Pi-ta fungal pathosystem in rice (Orbach et al., 2000). In the current study, we found that Avr-Pita suppresses chitin-triggered ROS accumulation and PR gene expression (Figs. 1d–e, 5a and S1f) and enhances susceptibility to M. oryzae in rice (Fig. 1a–c).
Pathogen effectors can be divided into apoplastic and intracellular effectors based on their localization. Most bacterial effectors and some fungal effectors are dispersed in the apoplastic space, such as the M. oryzae effector Slp1 (Mentlak et al., 2012), the Ustilago maydis effectors Pep1 (Doehlemann et al., 2009) and Pit2 (Mueller et al., 2013), and the Cladosporium fulvum effectors Avr2 (Rooney et al., 2005) and Avr4 (van den Burg et al., 2006). Intracellular effectors are secreted by invasive hyphae and delivered into host cells. The fungal effectors PWL1 and PWL2 (Khang et al., 2010), AvrPiz-t (Park et al., 2012), and Avr-Pii (Singh et al., 2016) initially accumulate in the BIC structure. After they enter the host cells, PWL2 (Khang et al., 2010), AvrPiz-t (Park et al., 2012), and Avr1-CO39 (Ribot et al., 2013) localize to the cytoplasm, and Avr-Pib partially targets the host nucleus (Zhang et al., 2018). Avr-Pita also accumulates in the BIC structure of M. oryzae during rice cell invasion (Khang et al., 2010). In the current study, we demonstrated that Avr-Pita targets the rice mitochondria (Fig. 3a and b, S3a and S3b) and interacts with OsCOX11 to regulate mitochondrial COX activity (Fig. 2a and b). Avr-Pita specifically binds to the region of 2th β sheet to 5th β sheet of the OsCOX11. Especially, the 5th β sheet is critical for the interaction, indicating that Avr-Pita may modify the specific residues in 5th β sheet of OsCOX11 as a metalloprotease to promotes the COX activity. The heterozygous oscox11/OsCOX11 knockout mutants, in which the OsCOX11 expression reduced ~ 50%, become resistant to the fungal pathogen (Fig. 4d and e). However, the sensitivity of current method for COX activity measurement may be too low to detect the expected COX activity reduction in the oscox11/OsCOX11 plants (Fig. S4c).
ROS such as H2O2, O2·−, and 1O2 play critical roles in plant immunity. In response to the perception of pathogen invasion, plants activate a rapid ROS burst and induce the HR via an immune signaling cascade (Kou et al., 2019). Apoplastic ROS production mediated by plasma membrane NADPH oxidases or cell wall peroxidases provides a major source of ROS for plant defense responses (Kadota et al., 2015). During plant–pathogen interactions, pathogens secrete a number of effectors into the host apoplast and cytoplasm. These apoplastic effectors interfere with PAMP perception mediated by plant membrane-bound pattern recognition receptor (PPR), leading to the inactivation of NADPH oxidases. By contrast, cytoplasmic effectors target the MAPK signaling pathway, vesicle trafficking, and metabolic priming, which are essential for apoplastic ROS production (Jwa & Hwang, 2017). Chloroplast-generated ROS also contribute to plant defense responses. The photosynthetic electron transport chains of photosystem I (PSI) and photosystem II (PSII) are two main sources of ROS production in chloroplasts (Lu & Yao, 2018). Several bacterial effectors such as HopNI and HopK1 target the chloroplast and manipulate chloroplast function associated with ROS production (Kretschmer et al., 2019). The fungal effector ToxA induces plant ROS accumulation in chloroplasts, which leads to cell death (Manning et al., 2009). The accumulation of ROS is light dependent and is associated with the dysfunction of the PSI and PSII complexes (Manning et al., 2009).
Mitochondria are important organelles for energy generation and ROS production (Jacoby et al., 2012; Mackenzie & McIntosh, 1999). Mitochondrial ROS, a major source of ROS in plant cells, is produced by Complexes I, II, and III of the METC, while the terminal complex (Complex IV) functions in ROS consumption via the COX-mediated transfer of electrons to molecular oxygen, eventually forming H2O (Timon-Gomez et al., 2018). Mitochondrial oxidase of the respiratory chain is a determinant of ROS generation involved in pathogen defense responses (Cvetkovska & Vanlerberghe, 2013; Vidal et al., 2007). The cytoplasmic male sterility protein WA352 interacts with OsCOX11 and inhibits its function in ROS scavenging, thereby inducing tapetal PCD in rice (Luo et al., 2013). Decreasing AtCOX11 expression suppresses COX activity in Arabidopsis, resulting in abnormal plant growth and pollen germination (Radin et al., 2015). However, no pathogen effectors have been shown to target mitochondria and interfere with mitochondrial ROS metabolism. In the current study, we demonstrated that silencing OsCOX11 limits pathogen propagation (Fig. 4d–f and S4e–S4g), whereas overexpressing this gene inhibits ROS accumulation and enhances susceptibility to M. oryzae (Figs. 4a–c and 5a–c), indicating that COX-mediated ROS metabolism is involved in plant resistance to this fungus. Furthermore, overexpressing Avr-Pita increased COX activity (Fig. 6) and suppressed ROS accumulation (Fig. 5) in rice, suggesting that the pathogen delivers the effector Avr-Pita to suppress host immune responses by enhancing OsCOX11-dependent ROS metabolism (Fig. 7).
Conclusions
Pathogens produce effectors to hinder host defenses, such as defense gene expression, or the production of ROS, which act as signaling molecules and (at higher concentrations) can directly kill pathogens. Avr-Pita was the first fungal effector protein identified from M. oryzae, the devastating fungal pathogen that causes rice blast disease, but its role in this plant–pathogen interaction is poorly understood. In current study, we provide evidence that Avr-Pita targets rice mitochondria and interacts with the COX assembly protein OsCOX11 of the METC. This interaction promotes the activity of COX to decrease ROS accumulation, thereby suppressing innate immunity in rice. This novel finding increases our understanding of how a pathogen effector dampens host innate immunity by hijacking a plant protein and manipulating mitochondrial activity in the M. oryzae–rice interaction.
Material and Methods
Generation and Characterization of Transgenic Plants
To generate transgenic rice lines with estradiol-inducible expression of Avr-Pita, a truncated fragment Avr-Pita145–672 encoding a mature protease was isolated and subcloned into the estradiol-inducible binary expression vector pXVE. The resulting construct (PXVE::Avr-Pita) was transformed into rice variety Kinmaze (without Pi-ta). To generate transgenic rice plants with stable overexpression of Avr-Pita or OsCOX11, the Avr-Pita145–672 fragment and full-length coding sequence of OsCOX11 were individually inserted into the binary vector pOX (containing the maize Ubiquitin promoter) to generate the PUbi::Avr-Pita and PUbi::OsCOX11 constructs, which were introduced into the rice cultivar Nipponbare (without Pi-ta).
Knockout of OsCOX11 in rice cultivar Nipponbare was performed by CRISPR/Cas9 gene editing as described previously (Ma et al., 2015). In brief, the 20-bp target site of OsCOX11 (5′-TTTAATGCTGACGTTGCTGA-3′) driven by the U6b promoter was cloned into the binary vector pYLCRISPR-Cas9Pubi-H. To silence the expression of OsCOX11 in the rice cultivar Tohoku IL6 background (carrying Pi-ta), ~ 0.35-kb sense and antisense fragments of the OsCOX11 coding sequence were cloned into the binary vector POsCERK1-RNAi (driven by the OsCERK1 promoter) to produce the POsCERK1::OsCOX11-RNAi construct.
The genotypes of OsCOX11 knockout plants were investigated by PCR using primer pair OsCOX11-In2Ex4-F/OsCOX11-In2Ex4-R, followed by sequencing with the OsCOX11-seq-F primer (Table S2). To measure Avr-Pita and OsCOX11 transcript levels in the transgenic plants, leaf blades were sampled from T0 or T1 plants and immediately frozen in liquid nitrogen for RNA extraction. Gene expression was then measured as described below.
Pathogen Inoculation and Evaluation of Disease Resistance
M. oryzae isolates 10–441 and 13–219, which are virulent on rice varieties Nipponbare and Tohoku IL6, were used to assess the resistance of rice to blast disease. The isolates were grown on Potato Dextrose Agar medium, and conidial production was induced on Oatmeal Agar medium under 12-h dark/12-h blue-light irradiation.
For leaf spray inoculation, three- to four-week-old seedlings were spray-inoculated with spore suspension (0.5 × 105 spores/mL, containing 0.01% Tween 20). The inoculated plants were maintained in a growth chamber at 26 °C with 90% humidity in the dark for 24 h, followed by incubation under a 12 h light/12 h dark cycle. The inoculated leaves were sampled at 0, 6, 12, 24, and 48 h for gene expression analysis.
For leaf punch inoculation, the young leaves of six- to eight-week-old plants were wounded with a mouse ear puncher. A piece of a M. oryzae spore colony was affixed to the wounding site and sealed with transparent tape. The inoculated leaves were collected and photographed at 12 dpi. The size of the lesion area was quantified using Image J software. Relative fungal biomass in infected leaves was measured by DNA-based quantitative PCR using MoPot2- and OsUbq-specific primers (Table S2) and determined using the eq. 2[CT(OsUbq)-CT(MoPot2)] (Berruyer et al., 2006).
To inoculate rice leaf sheaths, ~ 5-cm leaf sheath samples were collected from four- to six-week-old plants and injected with fungal spores (0.5 × 105 spores/mL, in 0.2% gelatin) in their hollow interiors. The infected samples were incubated in a 90% humidity chamber at 26 °C in the dark, and ROS detection was conducted at 0, 24, 48, and 96 h.
Gene Expression Analysis
To investigate the expression levels of defense response genes in different transgenic lines, leaf discs or suspension cells were treated with 200 μg/mL chitin and sampled at 0, 3, and 6 h. Suspensions cells derived from PXVE::Avr-Pita transgenic lines were treated with estradiol and sampled at 0, 3, and 6 h for expression analysis.
Total RNA was extracted from various rice tissues using TRIzol reagent (Thermo Fisher Scientific), and gDNA was removed using RNase-free RQ1 DNaseI (Promega). First-strand cDNA was synthesized with oligonucleotide dT primers and M-MLV reverse transcriptase (Promega). Transcript levels were quantified by qRT-PCR with gene-specific primers (Table S2) using SYBR Green master mix (Bio-Rad) on a CFX96 Real-Time System (Bio-Rad). The relative expression level of each gene was normalized to that of OsACTIN using the 2−ΔCt method.
Yeast Two-Hybrid Assays
The interactions between Avr-Pita and various COX11 truncated proteins were analyzed using the GAL4 Y2H system (Clontech). The Avr-Pita145–672 fragment was cloned into bait vector pGBKT7 (BD). Full-length fragments of the OsCOX11, AtCOX11, ScCOX11, and MoCOX11 coding sequences were amplified from the corresponding organisms and subcloned separately into the prey vector pGADT7 (AD). A series of truncated OsCOX11 prey vectors including OsCOX111–198, OsCOX1180–220, OsCOX11110–220, and OsCOX11140–244 were generated based on the OsCOX11 prey vector using the Ω-PCR method (Chen et al., 2013).
Various combinations of the bait and prey constructs were co-transformed into yeast Y2H Gold cells using the Matchmaker Gold Yeast Two-Hybrid System (Clontech). The transformants were grown on synthetic dropout medium without Leucine and Tryptophan (SD-LW) and synthetic dropout medium lacking Adenine, Histidine, Leucine, and Tryptophan (SD-AHLW) at 30 °C for 3 days.
Pull-Down Assays
The GST:Avr-Pita and His:OsCOX11 plasmids used for the pull-down assay were generated using the Ω-PCR method. Briefly, the Avr-Pita145–672 fragment and full-length OsCOX11 were individually subcloned into pGEX-4 T-2 (containing GST tag) (Sigma) and the prokaryotic expression vector pET32a (containing N-Trx, N-6×His, N-thrombin, N-S, N-enterokinase, C-6×His tag) (Novagen) by Ω-PCR (Chen et al., 2013).
The fusion proteins were expressed in E. coli BL21 cells, purified using GST or HIS binding resin, and subjected to pull-down assays. The pulled-down proteins were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Specific anti-GST and anti-His antibodies and HRP-conjugated antibody were used for immunoblot analysis. The signals were visualized with Immobilon Western HRP substrate and detected by automatic chemiluminescence image analysis (Tanon 5200 Multi).
Subcellular Localization of Fusion Proteins
Subcellular localization analysis of various fusion proteins was performed using transient expression vectors Avr-Pita:YFP and OsCOX11:mCherry carrying the Avr-Pita145–672 fragment and full-length cDNA of OsCOX11, respectively, driven by the 35S promoter.
The subcellular localization of Avr-Pita:YFP and OsCOX11:mCherry was investigated in rice protoplasts or onion epidermal cells. For transient expression in rice protoplasts, the indicated plasmids were co-transfected into rice protoplasts using PEG/Ca-mediated transfection and incubated for 12–18 h at 28 °C in the dark. The plasmids were also introduced into onion epidermal cells using a DNA particle delivery system (Bio-Rad, Helios Gene Gun). Following bombardment, the onion epidermal cells were incubated on Murashige and Skoog medium for 16–20 h at 28 °C in the dark. The transfected rice protoplasts or onion epidermal cells were observed under a laser-scanning confocal microscope (Carl Zeiss, LSCM 7DUO) with excitation laser wavelengths of 514 nm (YFP) and 543 nm (mCherry).
Detection of Reactive Oxygen Species
To measure ROS accumulation using the luminol-chemiluminescence method, rice suspension cell cultures containing 50 μM L-012, 2 mg/mL horseradish peroxidase, and 20 mg/mL chitin ((GlcNAc)8) (distilled water was used as a control) were introduced into 96-well plates. Luminescence was immediately measured at 30-s intervals for 120 min using a Multi-Mode Microplate Reader (BioTek, SynergyMx). Four biological replications were performed for each sample.
To detect the ROS using DAB staining, fungal-infected rice leaf sheaths were submerged in 1 mg/mL DAB solution (pH 3.8), and chlorophyll was completely removed from the samples with 95% boiling ethanol. The DAB-stained ROS appeared reddish-brown. The stained leaf sheaths were photographed under an optical microscope (Carl Zeiss, Axio Observer Z1).
Measuring Cytochrome c Oxidase Activity
Mitochondrial protein was extracted from rice tissues for COX activity measurements using a Mitochondrial Complexes IV Assay Kit (Suzhou Keming biotechnology) according to the manufacturer’s protocol. Briefly, chitin-treated (200 μg/mL) rice tissues were ground in liquid nitrogen and homogenized in ice-cold Extraction Buffer containing protease inhibitor. The supernatants were harvested by centrifugation, and COX activity in each sample was measured with a Multi-Mode Microplate Reader (BioTek, SynergyMx).
Availability of Data and Materials
All relevant data are provided within the article and its supplementary information files.
Abbreviations
- Avr:
-
Avirulence
- BIC:
-
Biotrophic interfacial complex
- COX:
-
Cytochrome c oxidase
- ETI:
-
Effector-triggered immunity
- HR:
-
Hypersensitive response
- MAPK:
-
Mitogen activated protein kinase
- METC:
-
Mitochondrial electron transport chain
- NBS-LRR:
-
Nucleotide binding site-leucine rich repeat domain
- PAMP:
-
Pathogen-associated molecular pattern
- PCD:
-
Programmed cell death
- PR:
-
Pathogenesis-related
- PTI:
-
PAMP-triggered immunity
- PSI:
-
Photosystem I
- PSII:
-
Photosystem II
- ROS:
-
Reactive oxygen species
References
Berruyer R, Poussier S, Kankanala P, Mosquera G, Valent B (2006) Quantitative and qualitative influence of inoculation methods on in planta growth of rice blast fungus. Phytopathology 96:346–355
Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8:521–539
Bryan GT, Wu K, Farrall L, Jia Y, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12:2033–2045
Chen L, Wang F, Wang X, Liu Y (2013) Robust one-tube Ω-PCR strategy accelerates precise sequence modification of plasmids for functional genomics. Plant Cell Physiol 54:634–642
Cvetkovska M, Vanlerberghe GC (2013) Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ 36:721–732
Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430
Deslandes L, Rivas S (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci 17:644–655
Doehlemann G, van der Linde K, Assmann D, Schwammbach D, Hof A, Mohanty A, Jackson D, Kahmann R (2009) Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog 5:e1000290
Fujisaki K, Abe Y, Ito A, Saitoh H, Yoshida K, Kanzaki H, Kanzaki E, Utsushi H, Yamashita T, Kamoun S, Terauchi R (2015) Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J 83:875–887
Jacoby RP, Li L, Huang S, Pong LC, Millar AH, Taylor NL (2012) Mitochondrial composition, function and stress response in plants. J Integr Plant Biol 54:887–906
Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19:4004–4014
Jwa NS, Hwang BK (2017) Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front Plant Sci 8:1687
Kadota Y, Shirasu K, Zipfel C (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol 56:1472–1480
Khang CH, Berruyer R, Giraldo MC, Kankanala P, Park S, Czymmek K, Kang S, Valent B (2010) Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22:1388–1403
Kou Y, Qiu J, Tao Z (2019) Every coin has two sides: reactive oxygen species during rice Magnaporthe oryzae interaction. Int J Mol Sci 20:1191
Kretschmer M, Damoo D, Djamei A, Kronstad J (2019) Chloroplasts and plant immunity: where are the fungal effectors? Pathogens 9:19
Liu W, Liu J, Triplett L, Leach JE, Wang G (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol 52:213–241
Lu Y, Yao J (2018) Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int J Mol Sci 19:3900
Luo D, Xu H, Liu Z, Guo J, Li H, Chen L, Fang C, Zhang Q, Bai M, Yao N, Wu H, Wu H, Ji C, Zheng H, Chen Y, Ye S, Li X, Zhao X, Li R, Liu Y (2013) A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet 45:573–577
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Mackenzie S, McIntosh L (1999) Higher plant mitochondria. Plant Cell 11:571–586
Manning VA, Chu AL, Steeves JE, Wolpert TJ, Ciuffetti LM (2009) A host-selective toxin of Pyrenophora tritici-repentis, Ptr ToxA, induces photosystem changes and reactive oxygen species accumulation in sensitive wheat. Mol Plant Microbe Interact 22:665–676
Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, Saitoh H, Terauchi R, Nishizawa Y, Shibuya N, Thomma BP, Talbot NJ (2012) Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24:322–335
Mueller AN, Ziemann S, Treitschke S, Assmann D, Doehlemann G (2013) Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog 9:e1003177
Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12:2019–2032
Park C, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M, Valent B, Wang G (2012) The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern–triggered immunity in rice. Plant Cell 24:4748–4762
Park CH, Shirsekar G, Bellizzi M, Chen S, Songkumarn P, Xie X, Shi X, Ning Y, Zhou B, Suttiviriya P, Wang M, Umemura K, Wang GL (2016) The E3 ligase APIP10 connects the effector AvrPiz-t to the NLR receptor Piz-t in rice. PLoS Pathog 12:e1005529
Qi J, Song C, Wang B, Zhou J, Kangasjarvi J, Zhu J, Gong Z (2018) Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol 60:805–826
Radin I, Mansilla N, Rodel G, Steinebrunner I (2015) The Arabidopsis COX11 homolog is essential for cytochrome c oxidase activity. Front Plant Sci 6:1091
Ribot C, Cesari S, Abidi I, Chalvon V, Bournaud C, Vallet J, Lebrun MH, Morel JB, Kroj T (2013) The Magnaporthe oryzae effector AVR1-CO39 is translocated into rice cells independently of a fungal-derived machinery. Plant J 74:1–12
Rooney HC, Van’T KJ, van der Hoorn RA, Joosten MH, Jones JD, de Wit PJ (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308:1783–1786
Shi X, Long Y, He F, Zhang C, Wang R, Zhang T, Wu W, Hao Z, Wang Y, Wang GL, Ning Y (2018) The fungal pathogen Magnaporthe oryzae suppresses innate immunity by modulating a host potassium channel. PLoS Pathog 14:e1006878
Singh R, Dangol S, Chen Y, Choi J, Cho YS, Lee JE, Choi MO, Jwa NS (2016) Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol Cells 39:426–438
Skamnioti P, Gurr SJ (2009) Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol 27:141–150
Smirnoff N, Arnaud D (2019) Hydrogen peroxide metabolism and functions in plants. New Phytol 221:1197–1214
Tang M, Ning Y, Shu X, Dong B, Zhang H, Wu D, Wang H, Wang G, Zhou B (2017) The Nup98 homolog APIP12 targeted by the effector AvrPiz-t is involved in rice basal resistance against Magnaporthe oryzae. Rice 10:5
Timon-Gomez A, Nyvltova E, Abriata LA, Vila AJ, Hosler J, Barrientos A (2018) Mitochondrial cytochrome c oxidase biogenesis: recent developments. Semin Cell Dev Biol 76:163–178
van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, de Wit PJ (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19:1420–1430
Vanlerberghe GC (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14:6805–6847
Vidal G, Ribas-Carbo M, Garmier M, Dubertret G, Rasmusson AG, Mathieu C, Foyer CH, De Paepe R (2007) Lack of respiratory chain complex I impairs alternative oxidase engagement and modulates redox signaling during elicitor-induced cell death in tobacco. Plant Cell 19:640–655
Wang R, Ning Y, Shi X, He F, Zhang C, Fan J, Jiang N, Zhang Y, Zhang T, Hu Y, Bellizzi M, Wang G (2016) Immunity to rice blast disease by suppression of effector-triggered necrosis. Curr Biol 26:2399–2411
Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D, Long Y, Zhong Y, Liu YG (2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18:676–687
Wilson RA, Talbot NJ (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 7:185–195
Zhang S, Xu J (2014) Effectors and effector delivery in Magnaporthe oryzae. PLoS Pathog 10:e1003826
Zhang X, He D, Zhao Y, Cheng X, Zhao W, Taylor IA, Yang J, Liu J, Peng Y (2018) A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus. Plant J 96:133–146
Acknowledgements
We dedicate this paper to Prof. Ko Shimamoto of Nara Institute of Science and Technology, Japan, who passed away and kindly provided original materials for this study.
Funding
This work was funded by grants from the National Key Research and Development Program of China (2016YFD0100601), the National Natural Science Foundation of China (31772104, 31071646, 32001434), and the Natural Science Foundation of Guangdong Province (2018B030311035).
Author information
Authors and Affiliations
Contributions
LC, JS and JH designed the research; JH, XW, FW, ZZ and GL performed experiments; JH, XW, FW and XW analyzed data; LC, JS and JH wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for Publication
Not applicable.
Competing interests
The authors declare that no competing interests exist.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Figure S1 Ectopic expression of Avr-Pita in rice. (a) Diagram of the PXVE::Avr-Pita and PUbi::Avr-Pita constructs. In these constructs, a truncated Avr-Pita145–672 fragment encoding mature protease is driven by the estradiol-inducible promoter XVE or the maize Ubiquitin (Ubi) promoter. SP: signal peptide; Pro: predicted prosequence. (b) The growth and developmental morphology of PUbi::Avr-Pita plants were not obviously affected in these lines. Scale bars: 10 cm. (c) Transcript levels of Avr-Pita was measured by RT-PCR in PXVE::Avr-Pita transgenic suspension cell lines after estradiol treatment. (d) Transcript levels of Avr-Pita was measured by RT-PCR in PUbi::Avr-Pita transgenic lines. (e) The expression of Avr-Pita in PXVE::Avr-Pita transgenic suspension cell lines after estradiol treatment using qRT-PCR. (f) The expression of defense-response gene OsPAL1 in PXVE::Avr-Pita suspension cell lines after estradiol treatment using qRT-PCR. (g) Overexpression of Avr-Pita in PUbi::Avr-Pita transgenic lines using qRT-PCR. OsACTIN served as a control to normalize the expression levels of target genes. Data are shown as mean ± SD (**P < 0.01, n = 3). Figure S2 Avr-Pita specifically binds to the conserved domains of OsCOX11. (a) Amino acid sequence alignment of COX11 orthologs OsCOX11 (O. sativa, XP_006650503.1), AtCOX11 (A. thaliana, AAG00893), ScCOX11 (S. cerevisiae, NP_015193), and MoCOX11 (M. oryzae, XP_003717808). Six β sheets (indicated by a single underline and numbered 1–6) and Cu-binding core region CFCF (indicated by four triangles ▲) are present in the conserved region of COX11. Residues 140–220 of OsCOX11, which are responsible for the interaction with Avr-Pita, are double underlined. The critical region OsCOX11199–220 is labelled with boxes. (b) Avr-Pita specifically interacts with rice OsCOX11 in a Y2H assay. Yeast cells were cultured on selective medium SD-LW or SD-AHLW; cell growth on SD-AHLW indicates a positive interaction. Figure S3 Avr-Pita and OsCOX11 co-localize to the mitochondria in onion epidermal cells. (a) Avr-Pita:YFP was introduced into onion epidermal cells by particle bombardment and stained with the mitochondrial dye MitoTracker. (b) Avr-Pita:YFP and OsCOX11:mCherry were transiently introduced into onion epidermal cells by particle bombardment. Scale bar = 50 μm. Figure S4 Characterization of OsCOX11 transgenic plants and pathogen resistance of OsCOX11-RNAi plants. (a) Expression levels of OsCOX11 in PUbi::OsCOX11 lines; OsACTIN served as an internal control. Data are shown as mean ± SD (**P < 0.01, n = 3). (b) Genotypes of heterozygous oscox11/OsCOX11 lines carrying a “T” or “C” base insertion. (c) Expression levels of OsCOX11 in oscox11/OsCOX11 plants. OsACTIN served as an internal control. Data are shown as mean ± SD (**P < 0.01, n = 3). (d) Expression levels of OsCOX11 in OsCOX11-RNAi plants. OsACTIN served as an internal control. Data are shown as mean ± SD (**P < 0.01, n = 3). (e) Disease symptoms of OsCOX11-RNAi transgenic plants at 12 dpi inoculated with M. oryzae isolate 13–219. (f) Lesion area in OsCOX11-RNAi transgenic plants at 12 dpi inoculated with M. oryzae isolate 13–219. Data are shown as mean ± SD (**P < 0.01, n > 12). (g) Relative fungal biomass on inoculated leaves at 12 dpi, as determined by qPCR. Data are shown as mean ± SD (**P < 0.01, n = 3). Figure S5 OsCOX11 expression in response to chitin and M. oryzae treatment. (a) The expression levels of OsCOX11 in PUbi::Avr-Pita and WT plants after chitin treatment. (b) The expression levels of OsCOX11 in rice seedlings in response to M. oryzae compatible strain 08-T13 inoculation at the indicated time points. OsACTIN served as an internal control. Data are shown as mean ± SD (**P < 0.01, n = 3). Table S1 CCandidates of Avr-Pita interacting protein screened by Y2H. Table S2 Primers used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Han, J., Wang, X., Wang, F. et al. The Fungal Effector Avr-Pita Suppresses Innate Immunity by Increasing COX Activity in Rice Mitochondria. Rice 14, 12 (2021). https://doi.org/10.1186/s12284-021-00453-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-021-00453-4