Abstract
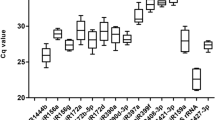
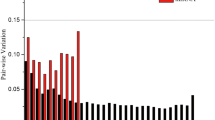
The microRNAs role in various cellular and metabolic functions is gaining more limelight in line with second-generation NGS technology. For the validation of candidate miRNA genes, the quantitative real-time PCR is the widely trusted and efficient method to follow. Sugarcane miRNAs are less explored in sugarcane defense response during their interaction with Colletotrichum falcatum inciting red rot. Further, for RT-qPCR experiments involving sugarcane miRNA expression studies, a stable internal reference gene is required. Hence, we have taken a study involving 20 candidate genes to identify stable expressing reference genes using NormFinder, geNorm, BestKeeper, and deltaCt statistical algorithms. The candidate reference genes included miRNAs and protein-coding genes. The results indicated that there is a variation in ranking among the algorithms. We found miR1862c as the stably expressed miRNA reference gene among the candidates and miR444b.2 along miR1862c formed the best reference gene pair combination, which can be used in the experiments aiming to explore sugarcane miRNAs in the defense mechanism against C. falcatum. The stable miRNA reference gene was further validated with other lesser stable reference gene candidates to assess the effect of stable reference genes during normalization. The present study evaluating the sugarcane miRNAs as reference genes for normalizing RT-qPCR expression data involving miRNAs during sugarcane × C. falcatum interaction is the first of its kind. Further, this systematic approach can be followed to assess the reference gene in various experimental conditions involving sugarcane miRNAs.



Similar content being viewed by others
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR Data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Ashwin NMR, Barnabas EL, Sundar AR et al (2017) Disease suppressive effects of resistance-inducing agents against red rot of sugarcane. Euro J Plant Pathol 149:285–297
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Bottino CM, Rosario S, Grativol C et al (2013) High-throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS ONE 8:e59423
Brodersen P, Voinnet O (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10:141–148. https://doi.org/10.1038/nrm2619
Busk PK (2014) A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinform. https://doi.org/10.1186/1471-2105-15-29
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25(2):169–193
Bustin SA, Benes V, Garson JA et al (2009) The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
de Andrade LM, dos Santos BM, Fávero Peixoto Junior R et al (2017) Reference genes for normalization of qPCR assays in sugarcane plants under water deficit. Plant Methods. https://doi.org/10.1186/s13007-017-0178-2
de Silva RLO, Silva MD, Ferreira JRCN et al (2014) Validation of novel reference genes for reverse transcription quantitative real-time PCR in drought-stressed sugarcane. Sci World J 2014:1–12. https://doi.org/10.1155/2014/357052
Derveaux S, Vandesompele J, Hellemans J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50:227–230
Die JV, Román B, Nadal S, González-Verdejo CI (2010) Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 232(145–153):1
Fausto AKS, da Silva TF, Romanel E, Vaslin MFS (2017) microRNAs as reference genes for quantitative PCR in cotton. PLoS ONE 12:e0174722
Ferreira TH, Gentile A, Vilela RD et al (2012) microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS ONE 7:e46703
Ganesh VK, Viswanathan R, Malathi P et al (2020) Identification of differential expressed proteins and establishing a defense proteome of sugarcane in response to Colletotrichum falcatum infection. J Plant Pathol. https://doi.org/10.1007/s42161-020-00577-4
Guo J, Ling H, Wu Q et al (2014) The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci Rep. https://doi.org/10.1038/srep07042
Iskandar HM, Simpson RS, Casu RE et al (2004) Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol Biol Rep 22:325–337
Kulcheski FR, Marcelino-Guimaraes FC, Nepomuceno AL et al (2010) The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal Biochem 406:185–192
Kulcheski FR, de Oliveira LF, Molina LG et al (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. https://doi.org/10.1186/1471-2164-12-307
Ling H, Wu Q, Guo J et al (2014) Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS ONE 9:e97469. https://doi.org/10.1371/journal.pone.0097469
Ling H, Huang N, Xu L et al (2019) Suitable reference genes/miRNAs for qRT-PCR normalization of expression analysis in sugarcane under Sorghum mosaic virus infection. Sugar Tech 21:780–793
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mentzel C, Skovgaard K, Cordoba S et al (2014) Wet-lab tested MicroRNA assays for qPCR studies with SYBR® green and DNA primers in pig tissues. MicroRNA 3:174–188
Mohanraj D, Padmanaban P, Viswanathan R (2012) Screening for red rot resistance in sugarcane. In: Viswanathan R, Sundar AR (eds) Functional plant science and biotechnology 6 (special issue 2). Global Science Books, Ikenobe, pp 51–62
Pfaffl MW, Tichopad A, Prgomet C et al (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper excel-based tool using pair-wise correlations. Biotech Lett 26:509–515
Rahul PR, Kumar VG, Sathyabhama M et al (2015) Characterization and 3D structure prediction of chitinase induced in sugarcane during pathogenesis of Colletotrichum falcatum. J Plant Biochem Biotechnol 24:1–8
Rahul PR, Ganesh Kumar V, Viswanathan R et al (2016) Defense transcriptome analysis of sugarcane and Colletotrichum falcatum interaction using host suspension cells and pathogen elicitor. Sugar Tech 18:16–28
Rhoades MW, Reinhart BJ, Lim LP et al (2002) Prediction of plant MicroRNA targets. Cell 110:513–520
Sathyabhama M, Viswanathan R, Nandakumar M et al (2015) Understanding sugarcane defence responses during the initial phase of Colletotrichum falcatum pathogenesis by suppression subtractive hybridization (SSH). Physiol Mol Plant Pathol 91:131–140
Sathyabhama M, Viswanathan R, Malathi P, Sundar AR (2016) Identification of differentially expressed genes in sugarcane during pathogenesis of Colletotrichum falcatum by suppression subtractive hybridization (SSH). Sugar Tech 18:176–183
Silver N, Best S, Jiang J, Thein S (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33. https://doi.org/10.1186/1471-2199-7-33
Song C, Fang J, Wang C et al (2010) MiR-RACE, a new efficient approach to determine the precise sequences of computationally identified trifoliate orange (Poncirus trifoliata) microRNAs. PLoS ONE 5:e10861
Sundara B (1998) Sugarcane cultivation. Vikas Publishing House, New Delhi
Thiebaut F, Rojas CA, Almeida KL et al (2012) Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ 35:502–512
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115–e115. https://doi.org/10.1093/nar/gks596
Vandesompele J, De Preter K, Pattyn F et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034. https://doi.org/10.1186/gb-2002-3-7-research0034
Viswanathan R (2010) Plant disease: red rot of sugarcane. Anmol Publications, New Delhi
Viswanathan R (2020) Sustainable sugarcane cultivation in India through threats of red rot by varietal management. Sugar Tech. https://doi.org/10.1007/s12355-020-00882-3
Viswanathan R, Selvakumar R (2020) Varietal breakdown to red rot in sugarcane revealed by comparing two Colletotrichum falcatum inoculation methods. Sugar Tech. https://doi.org/10.1007/s12355-020-00855-6
Viswanathan R, Sundar AR, Malathi P et al (2009) Interaction between sugarcane and Colletotrichum falcatum causing red rot: understanding disease resistance at transcription level. Sugar Tech 11:44–50
Viswanathan R, Sathyabhama M, Malathi P, Sundar AR (2016) Transcriptome analysis of host–pathogen interaction between sugarcane and Colletotrichum falcatum by suppression subtractive hybridization and Illumina sequencing. Proc Intern Soc Sugarcane Technol 29:1639–1644
Viswanathan R, Sundar AR, Selvakumar R, Malathi P (2018) Progress in understanding fungal diseases affecting sugarcane: red rot. In: Rott P (ed) Achieving sustainable cultivation of sugarcane breeding, pests and diseases. Burleigh Dodds Science Publishing, Cambridgeshire, pp 201–220
Viswanathan R, Padmanaban P, Selvakumar R (2020) Emergence of new pathogenic variants in Colletotrichum falcatum, stalk infecting ascomycete in sugarcane: role of host varieties. Sugar Tech 22:473–484
Yang Y, Zhang X, Chen Y et al (2016) Selection of reference genes for normalization of microRNA expression by RT-qPCR in sugarcane buds under cold stress. Front Plant Sci. https://doi.org/10.3389/fpls.2016.00086
Acknowledgements
The authors are grateful to the Director of the Institute for providing necessary facilities to carry out the work.
Funding
The part of the study received financial support from Sugarcane Development Fund (No. 6-7/2006-R&D/SDF/885), Ministry of Consumer Affairs, Food & Public, New Delhi.
Author information
Authors and Affiliations
Contributions
RV conceptualized, designed, initiated, and supervised the project. MN prepared materials and performed whole genome and transcriptome data analysis. MN performed the lab experiments and contributed to data analysis and data interpretations. RV, PM and ARS analyzed the interpreted whole information retrieved from genome and transcriptome. MN and RV wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
The present research did not involve human participants and/or animals.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Nandakumar, M., Viswanathan, R., Malathi, P. et al. Selection of reference genes for normalization of microRNA expression in sugarcane stalks during its interaction with Colletotrichum falcatum. 3 Biotech 11, 72 (2021). https://doi.org/10.1007/s13205-020-02632-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02632-4