Phylogenetic Relationships Within Chrysogorgia (Alcyonacea: Octocorallia), a Morphologically Diverse Genus of Octocoral, Revealed Using a Target Enrichment Approach
- 1CSIRO Oceans and Atmosphere, Hobart, TAS, Australia
- 2Discipline of Biological Sciences, School of Natural Sciences, University of Tasmania, Hobart, TAS, Australia
- 3Department of Biology, Harvey Mudd College, Claremont, CA, United States
- 4Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
- 5Littoral, Environnement et Sociétés, UMR 7266 CNRS, La Rochelle Université, La Rochelle, France
- 6Institut Systématique Evolution Biodiversité (ISYEB), Muséum national d’Histoire naturelle, CNRS, Sorbonne Université, Université des Antilles, EPHE, Paris, France
The octocoral genus Chrysogorgia (Duchassaing and Michelotti, 1864) contains 81 nominal species that are ecologically important components of benthic communities. Taxonomic examination of a large set of samples revealed many provisional new species, exhibiting a wide range of morphological variation. We established nine, distinct morphological groups of Chrysogorgia s.l. that were hypothesized to represent distinct genera. Here, we applied a recently developed universal target enrichment bait method for octocoral exons and ultraconserved elements (UCEs) on 96 specimens varying in morphology, collection ages and DNA quality and quantity to determine whether there was genetic support for these morphologically defined groups. Following Illumina sequencing and SPAdes assembly we recovered 1,682 of 1,700 targeted exon loci and 1,333 of 1,340 targeted UCE loci. Locus recovery per sample was highly variable and significantly correlated with time since specimen collection (2–60 years) and DNA quantity and quality. Phylogenetically informative sites in UCE and exon loci were ∼35% for 50% and 75% taxon-occupancy matrices. Maximum likelihood analyses recovered highly resolved trees with topologies supporting the recognition of 11 candidate genera, corresponding with morphological groups assigned a priori, nine of which are novel. Our results also demonstrate that this target-enrichment approach can be successfully applied to degraded museum specimens of up to 60 years old. This study shows that an integrative approach consisting of molecular and morphological methods will be essential to a proper revision of Chrysogorgia taxonomy and to understand regional diversity of these ecologically important corals.
Introduction
Octocorals, referred to colloquially as “soft corals” or “sea fans,” are non-reef forming, sessile invertebrates that are typically long-lived and slow growing (Roberts et al., 2009; Watling et al., 2011). These invertebrates are conspicuous components of benthic communities where they are ecologically important, providing habitat and refugia for other marine fauna (Mosher and Watling, 2009; Watling et al., 2011; De Clippele et al., 2019). Octocorals are also classified as “vulnerable marine fauna” and are protected from the destructive effects of bottom fishing in the high seas (FAO, 2009; Auster et al., 2011; Watling and Auster, 2017). The effective management and regulation of deep-sea marine environments necessitates knowledge about the biodiversity, distribution and abundance of octocorals (Ramirez-Llodra et al., 2010; Watling and Auster, 2017). However, historical difficulties with octocoral taxonomy hamper our ability to reliably identify species in this group during monitoring surveys, and consequently, define effective strategies for their conservation.
The genus Chrysogorgia (Duchassaing and Michelotti, 1864) (Family: Chrysogorgiidae Verrill, 1883), commonly known as “gold coral,” is one of the most abundant and diverse octocoral groups (Cairns, 2001), with 81 nominal species. They are distributed worldwide, occurring on hard and soft substrates, at depths of 31–4,327 m based on published records (Cairns, 2001, 2018; Pante et al., 2012) and our own assessment of museum specimens. The biogeography of Chrysogorgia has been investigated in both the south-western Pacific and north-western Atlantic oceans (Thoma et al., 2009; Pante et al., 2012, 2015). Several Chrysogorgia mitochondrial haplotypes, used as proxies for species, are found in both the Atlantic and Pacific oceans, suggesting either a high dispersal ability of these octocorals or ancient connections between ocean basins (Thoma et al., 2009; Pante et al., 2012, 2015). Ecologically, chrysogorgiids are host to a variety of associated invertebrates, including brittle stars (Mosher and Watling, 2009) and squat lobsters (Schnabel, 2020; Wicksten, 2020). Surveys in Tasmania, Australia, have found that fields of chrysogorgiids are common on historically fished seamounts, suggesting these corals maybe used as indicators of deep-sea community recovery or resilience (Althaus et al., 2009).
The taxonomy of Chrysogorgia has gained interest since the 2000s, partly due to an increase in deep-sea exploration (Pante and Watling, 2012). As such, several new species have been recently described (e.g., Pante and Watling, 2012; Cordeiro et al., 2015; Cairns, 2018; Xu et al., 2020) but a comprehensive taxonomic revision of the genus and a critical examination of morphological characters in nominal species of Chrysogorgia has not been conducted since Versluys (1902). Taxonomists have recognized the range of morphological variation in this genus and sought to categorize Chrysogorgia species into groups based on the nature of the sclerites found in the polyp body and tentacles (Wright and Studer, 1889; Versluys, 1902). Presently, three groups are recognized: Group A, the “Spiculosae,” consists of species that have rod/spindle shaped sclerites in the polyp body wall and tentacles, while Group C (“Squamosae”) species have scales in these regions. An intermediate, Group B (“Squamosae aberrantes”) have scales in the body wall and rods/spindles in the tentacles (Wright and Studer, 1889; Versluys, 1902; Cairns, 2001; Pante and Watling, 2012; Xu et al., 2020). Species within these constituent groups are then separated and defined based on additional morphological features including branching sequence, the angle between the main stem and branches and several other colony and polyp morphometrics as well as fine-scale sclerite features (e.g., Bayer and Stefani, 1988; Cairns, 2001, 2018; Pante and Watling, 2012).
An integrative approach, assessing both morphology and molecular variation, is needed to resolve the taxonomy of Chrysogorgia. Morphological characters are still required for formal taxonomic revision. However, species identification of octocorals relies on expert examination of fine-scale, microscopic features which are not easily interpreted or useable by non-experts. In addition, homoplasious morphological characters, subtle variation in morphology, and environmental plasticity can obscure species boundaries in octocorals (Ament-Velásquez et al., 2016; McFadden et al., 2017; Quattrini et al., 2019). While molecular methods have become a valuable tool in taxonomy, they can still exhibit deficiencies. For instance, studies of mitochondrial mtMutS divergence revealed conflict among variation within and between species of Chrysogorgia (Pante and Watling, 2012; Pante et al., 2015). Studies employing multiple molecular markers (e.g., mtMutS, mt-cox1, igr1, ND2 and nuclear 28s rDNA) are also not always effective at distinguishing octocoral species (McFadden et al., 2010, 2014a,b; Baco and Cairns, 2012; Quattrini et al., 2019). While molecular analyses of Chrysogorgiidae using mtMutS, 18S and mt-cox1 markers showed that Chrysogorgia appears monophyletic (Pante et al., 2012), and genetic variation in mtMutS is congruent with discrete morphologies that correspond to species boundaries in four Atlantic species (Pante and Watling, 2012), in each case only a small number of taxa (< 50) were surveyed and the need for more comprehensive DNA sequencing was identified (Pante and Watling, 2012; Pante et al., 2012). A much larger number of loci have been surveyed for Chrysogorgia using Restriction-site Associated DNA sequencing (RAD-seq) (Pante et al., 2015), however the phylogenetic resolution of RAD loci can drop severely with phylogenetic depth, a phenomenon known as locus dropout (e.g., Collins and Hrbek, 2018), the number of shared loci declining with phylogenetic divergence among taxa (Eaton et al., 2017). Also, this technique requires high quantity and quality DNA, which can generally only be extracted from relatively recently collected material (Faircloth et al., 2015; Leaché et al., 2015; Pante et al., 2015; Harvey et al., 2016). This inherently limits the use of old type and other museum material that may be fragmentary, fluid-preserved and/or dried and generally provide low quantity and quality DNA (McCormack et al., 2016).
Target-capture enrichment of ultra-conserved elements (UCEs) and exons has proven useful in capturing 100’s of loci from both dry and fluid-preserved museum material (McCormack et al., 2016; Ruane and Austin, 2017; Wood et al., 2018; Derkarabetian et al., 2019; Tsai et al., 2019). This technique offers advantages over traditional Sanger sequencing and other next-generation sequencing methods like RAD-seq because both UCE and exon loci distributed throughout the genome are captured and enriched by hybridizing short (∼120 bp) RNA sequences (i.e., probes) to sheared genomic DNA of each sample (Faircloth et al., 2012). Therefore, high molecular weight DNA from specimens is not essential (McCormack et al., 2016; Ruane and Austin, 2017; Wood et al., 2018; Derkarabetian et al., 2019; Tsai et al., 2019). Furthermore, UCEs have been used to study evolutionary relationships at various taxonomic levels (Faircloth, 2013; McCormack et al., 2016; Ruane and Austin, 2017; Wood et al., 2018), and thus can resolve phylogenies across deep to shallow timescales (Faircloth et al., 2013; Smith et al., 2014).
Target-capture enrichment of UCEs and exons was recently pioneered for anthozoans, resulting in the first comprehensive time-calibrated phylogeny with high support for subclass and ordinal-level relationships (Quattrini et al., 2018, 2020). This technique has subsequently been refined and used to investigate populations and species-level relationships in the diverse octocoral genera Alcyonium Linnaeus, 1775 and Sinularia May, 1898 (Erickson et al., 2020). These studies have shown that this technique has greater utility for resolving phylogenetic relationships and species boundaries in octocorals than existing single and multi-locus markers. However, this target enrichment approach has not been used on specimens collected more than 30 years ago and preserved in <95% EtOH for extended periods. Here, we test the utility of target-capture enrichment of UCEs and exons for analysis of museum specimens of Chrysogorgiidae. We then use these data to test congruence between assigned morphological groups and clades obtained in the phylogeny and also investigate biogeographic patterns across depth and ocean basin.
Materials and Methods
Taxon Sampling and Morphological Examination
Samples for DNA analysis were obtained from collections worldwide and comprised 88 specimens assigned to Chrysogorgia s.l., including six type specimens and 15 specimens assigned to known species. We also sampled eight outgroup specimens from other genera within Chrysogorgiidae. The sample set covered a range of geographic locations (Atlantic and Indo-Pacific oceans, between 50°S and 50°N; Figure 1), depths (62–3957 m), collection dates (1959–2018), collection methods and morphologies (Figure 1 and Supplementary Table 1). Taxonomic examination was conducted on a larger set of specimens (>600) and examination of 50 of the 81 valid Chrysogorgia type species. Thorough examination of morphological variation was conducted and included examination of established macroscopic characters such as colony form, branching sequence, orthostiche interval (defined as the distance between two reference branches in the branching sequence), branch lengths, polyp size and sclerite form and arrangement together with assignment to Versluys’ sclerite group (A, B, C) Versluys, 1902; Bayer and Stefani, 1988; Cairns, 2001; Pante and Watling, 2012). Microscopic examination of sclerite features were performed on several polyps within a colony, where material was available, by dissolving the tissue in 10% sodium hypochlorite and using light microscopy (Alderslade, 1998).
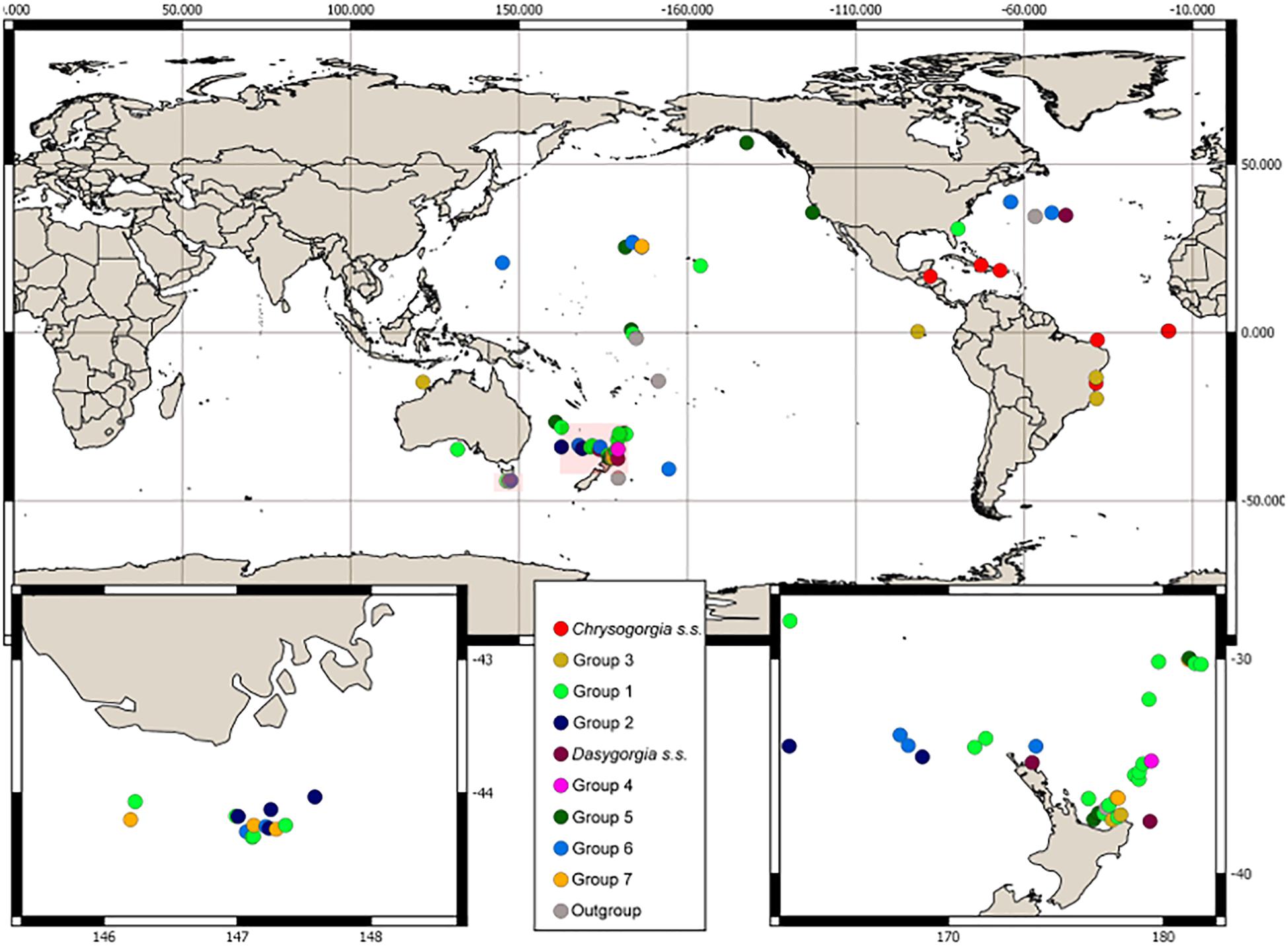
Figure 1. Collection location and morphological group assignment of samples. Locations of insets are southern Tasmania and northern New Zealand indicated by the pink boxes.
Molecular Procedures
DNA Extraction
Polyps were removed from the skeletal axis with a sterile scalpel and weighed after blotting excess ethanol, and then DNA extraction was conducted following a CTAB organic extraction (France et al., 1996). Polyps were macerated in 300 μL 2x CTAB buffer with a plastic pestle before adding 200 μg of Proteinase K and 200 μL of CTAB buffer. Samples were incubated at 65°C overnight, with an 200 μg of Proteinase K added after one hour. DNA was purified using three 500 μL organic extractions: first chloroform-isoamyl alcohol (24:1), second with phenol/chloroform-isoamyl alcohol (25:24:1),and finishing with chloroform-isoamyl alcohol (24:1). DNA was precipitated in 750 μL of cold isopropanol prior to centrifugation at 4°C, followed by rinsing the pellet with 400 μL of 70% ethanol to remove excess salt. DNA pellets were dried under vacuum and resuspended in 25 μL AE buffer. DNA quantity and quality were assessed with a Fragment Analyzer (Agilent 5200 Fragment Analyzer System). Before shipping to Arbor BioSciences, Ann Arbor, MI, United States, DNA extractions were stabilized by adding 6.25 μL of Biomatrica DNAstable® Plus.
Bait Set Design
An octocoral-specific bait set targeting exons and UCEs was used in the target enrichment, as described in Erickson et al. (2020). The bait set included 29,363 baits (120 bp) targeting 3,040 octocoral loci (1,340 UCE and 1,700 exon loci), with 3–18 baits targeting each locus. From this bait set, a subset (18,791 baits) designed only against species in the Calcaxonia-Pennatulacea clade (McFadden et al., 2006), to which Chrysogorgiidae belongs, were synthesized (Arbor BioSciences myBaits ®).
DNA Target Enrichment and Sequencing
Sample preparation, target enrichment and sequencing were performed through Arbor Biosciences as part of their myReads ®Targeted NGS services. DNA samples were sheared using a Qsonica Q800R sonicator and then size-selected to 500 bp with SPRI beads. Illumina Truseq-style, dual 8nt indexed libraries were constructed using either six or 10 cycles of indexing amplification following end repair and adapter ligation. A total of 22–167 ng of DNA was used per library (specimen), with six libraries pooled per enrichment reaction (16 enrichments), using the myBaits® kit and following the myBaits ® manual version 4.1, but substituting reagent Block A with IDT’s xGen universal blocking oligos, volume-for-volume. The myBaits ® were diluted to 1/2 (250 ng) of the standard reaction. Hybridizations were performed at 62°C for 23 h, with washes also performed at 62°C. Half of each captured product was re-amplified in 10 cycles using universal primers, quantified with library quantitative PCR, and then pooled in an equimolar concentration prior to sequencing. This pool was sequenced across 400M PE150 read-pairs on a NovaSeq S4 flowcell.
Postsequencing Analyses
Postsequencing analyses followed protocols in Quattrini et al. (2018) and PHYLUCE, following the workflow in the online tutorial I1, with a few modifications to the pipeline (Faircloth, 2016; Quattrini et al., 2018). Summary data and statistics were computed on all samples sequenced. Raw, untrimmed fastq read data were cleaned in order to remove adapter contamination and low quality bases using ILLUMIPROCESSOR with default values (Faircloth et al., 2013; Bolger et al., 2014), followed by contig assembly using SPAdes v. 3.10 (Bankevich et al., 2012). UCE and exon probes were separately matched to contigs (70% identity, 70% coverage) using phyluce_assembly_match_contigs_to_probes to find targeted loci. Both UCE and exon locus sets were extracted using phyluce_assembly_get_match_counts and relevant FASTA data were extracted using phyluce_assembly_get_fastas_from_match_counts. Files were aligned using phyluce_align_seqcap_align, which uses default settings in PHYLUCE. Data matrices of locus alignments were generated for each bait set using phyluce_align_get_only_loci_with_min_taxa where each locus had either 50% or 75% species occupancy. Concatenated locus alignments consisted of UCE, exon, or all loci. The number of parsimony informative sites was calculated for each 50% and 75% concatenated alignment matrix on unphased data using phyluce_align_get_informative_sites. The total number of variable sites, total number of parsimony informative sites and number of parsimony informative sites per locus were calculated using PHYLUCE (Faircloth, 2016).
Four samples (N025, N038, S080, and T060) had low locus recovery (< 120) for one or both probe sets and were removed from subsequent analysis (Supplementary Table 4). Two samples (C034, C040) were not included in the phylogeny as they were from multiple collections of the same colony as REM001D, and clustered with it in preliminary analyses. Existing fasta data for a specimen of Chrysogorgia tricaulis (ANT23) and an additional outgroup specimen, Radicipes (ANT176), from Quattrini et al. (2018), were added to make a final data set of 92 samples. Maximum Likelihood inference using RAxML v.8 (Stamatakis, 2014) was then conducted on 50% and 75% concatenated matrices for all loci combined. This analysis was carried out using rapid bootstrapping, which allows for a complete analysis (20 ML searches and 200 bootstrap replicates) in one step. Rooting was to outgroup genera: Metallogorgia (Versluys, 1902), Pseudochrysogorgia (Pante and France, 2010), Radicipes (Stearns, 1883), Rhodaniridogorgia (Watling, 2007) and Iridogorgia (Verrill, 1883). Biogeographic data were mapped onto the phylogeny in order to determine if there were patterns with depth and ocean basin. Depth distributions were expressed as ranges and classified into one of six categories according to collection event information: <500 m; 500–1000 m; 1000–1500 m; 1500–2000 m, 2000–3000 m, and >3000 m.
Recovery of mtMutS and 28S rDNA
Although not included in the bait set, we investigated the recovery of mtMutS and 28S rDNA sequences from our data. Assembled contigs were blasted against all available Chrysogorgia sequences from the NCBI database using ncbi blastn (e-value 0.01). Whole to partial mitochondrial genomes (including mtMutS) were uploaded to Mitos2 (Bernt et al., 2013) for annotation.
Results
Morphological Examination
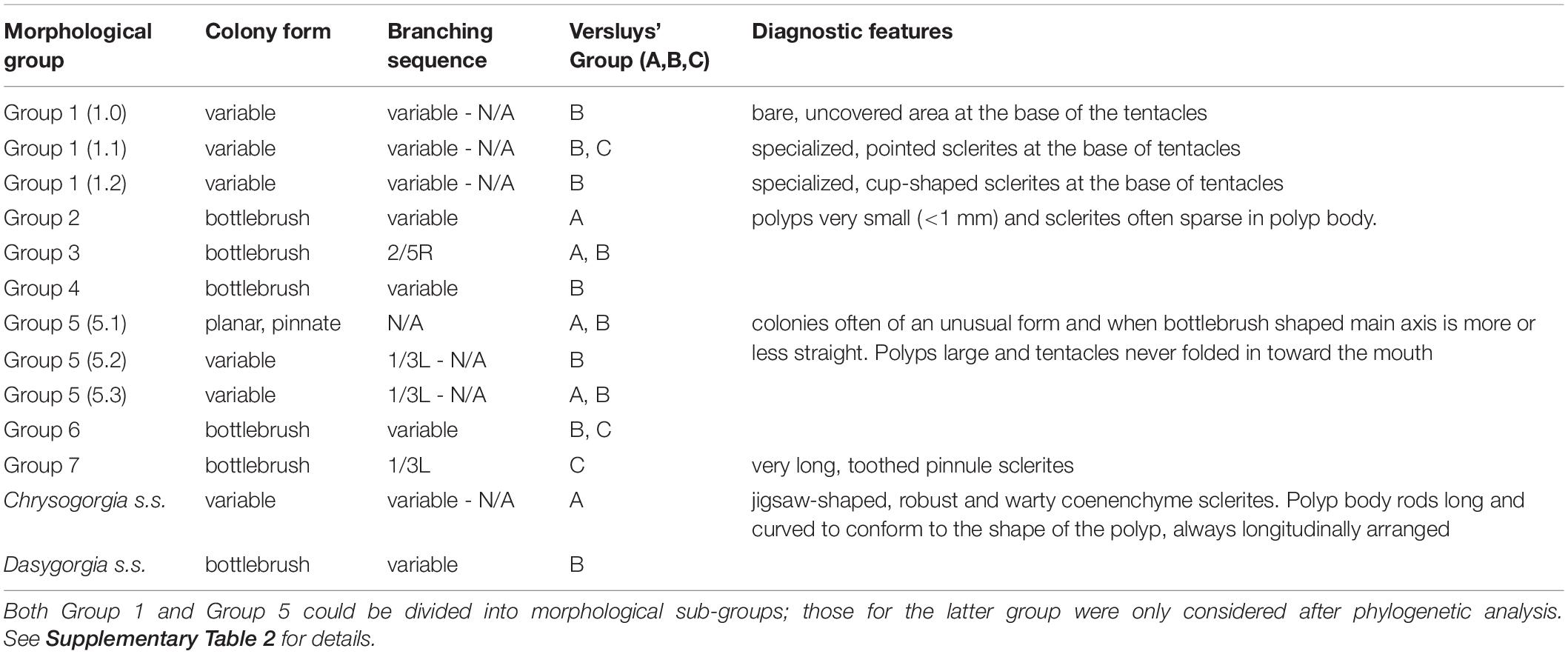
Following a detailed taxonomic investigation, 11 of the 15 specimens previously assigned to known species of Chrysogorgia were re-classified (Supplementary Table 2). An additional 16 specimens could be confidently assigned to known species, while 61 specimens were assigned to 53 putative new species. One of the outgroup specimens (OG09) was originally assigned to the genus Pseudochrysogorgia but re-assigned to Chrysogorgia following morphological identification (Supplementary Table 2). Given the wide range of morphological variation exhibited by specimens currently assigned to Chrysogorgia, we established nine, broad morphological groups and assigned specimens to them prior to our phylogenetic analysis (Table 1 and Supplementary Table 2). These morphological groups were based on an assessment of the following character set: colony form, branching sequence, polyp size, sclerite complement (expressed as Versluys’ group A, B, C), and presence of diagnostic features such as a bare, sclerite-free area at the base of the tentacles or specialized pinnule sclerites (Table 1).
Specimens assigned to Group 1 represent a large assemblage of species, which can adopt a variety of colony forms including bottlebrush-shaped as well as bi- and multi-flabellate forms. Branching sequence in the bottlebrush forms is variable and no branching sequence can be determined from bi- or multi-flabellate colonies, unless a significant portion of the single, main stem is present (Supplementary Table 2). Polyps of these specimens are very large in size (5–25 mm). Sclerites of the polyp body are always scales, which can be robust and take on a variety of shapes, sizes and arrangements. Sclerites in the basal part of the tentacle are either rods, often robust and warty or smooth, long, robust scales. This places species of this group into Versluys’ group B (“Squamosae abberantes”) or C (“Squamosae”). Differentiation from other groups is based on the presence of a bare, sclerite free area at the base of the tentacle, a character which is not present in any other group. Separation between species in this group is based on the shape and arrangement of sclerites directly underneath the bare area, which can be the same as those in the rest of the polyp body (designated as sub-group 1.0) or, more often, modified and arranged to form points (sub-group 1.1) or cups (sub-group 1.2) (Table 1 and Supplementary Table 2). Described species of Chrysogorgia s. l. in this group include C. octagonus (Versluys, 1902), C. calypso (Bayer and Stefani, 1988), C. bracteata (Bayer and Stefani, 1988), C. chryseis (Bayer and Stefani, 1988), C. stellata (Bayer and Stefani, 1988), C. versluysi (Kinoshita, 1913), C. electra (Bayer and Stefani, 1988), C. squamata (Verrill, 1883) and C. antarctica (Cairns, 2002) (Supplementary Table 3).
Group 2 represents specimens with a bottlebrush colony form having a 1/4L branching sequence, in most cases. Polyps in this group are very small (<1 mm), with sclerites often sparsely distributed in the polyp body wall of most polyps. Rods are also present in the tentacle backs and pinnule sclerites, when present, are not numerous. Species in this group therefore fall into the Spiculosae (Versluys’ group A), having a sclerite complement of rods only; these always have rounded ends and are small and uniformly sized throughout the colony (Table 1 and Supplementary Table 2). This group includes several Pacific and Indian Ocean species: C. papillosa (Kinoshita, 1913), C. rotunda (Kinoshita, 1913), C. dichotoma (Thomson and Henderson, 1906), and C. flexilis var. africana (Kükenthal, 1909) (Supplementary Table 3).
The morphological characters used to assign specimens to Group 3 were based on the taxonomic characters of some nominal species, specifically C. elegans (Verrill, 1883) and C. spiculosa (Verrill, 1883) (Supplementary Table 3). This group is characterized by a bottlebrush colony form with a regularly 2/5R branching sequence. Polyps are medium in size (3–8 mm) and sclerites in the polyp body are rods and/or spindles and occasionally also scales. The basal part of the tentacle has only rods and/or spindles. Sclerites are always densely packed and arranged obliquely longitudinally in the polyp body, modified in shape and size within a region in some species. This complement of sclerites places species assigned to this group in either Versluys’ group A or B (Table 1 and Supplementary Table 2).
Assignment of Group 4 was based on the taxonomic examination of the holotype of Chrysogorgia laevorsa (Cairns, 2018). The colony form is a small bottlebrush with variable branching sequence. The polyps are long (up to 20 mm) and slender, resembling those of Group 5, but with a sclerite complement of rods and spindles, as opposed to narrower, needle-shaped forms. Other species falling in this group are C. upsilonia (Cordeiro et al., 2015), C. tuberculata (Cordeiro et al., 2015), C. midas (Cairns, 2018) and C. debilis (Kükenthal, 1909). While this sclerite complement places these species in Versluys’ group A, many forms in either the polyp or tentacle region of some species can be markedly flattened and are thus considered as scales, which places them in Versluys’ sclerite group B.
Specimens were assigned to Group 5 based on the unusual form of their colonies and polyps with respect to others in Chrysogorgia s.l. The colony form in this group is generally planar with a pinnate branching pattern, and in specimens with a branched colony the main axis is more or less straight (Table 1). Polyps are large, narrow and elongate (up to 20 mm), with long tentacles that are always extended and never folded in toward the mouth. Polyp body sclerites can be scales, or more often, long, slender, needle-like forms; there are no rods. This group therefore falls in two Versluys sclerite groups A and B. Morphological Group 6 includes the nominal species C. abludo (Pante and Watling, 2012), C. dendritica (Xu et al., 2020) and C. pinnata (Cairns, 2001) (Supplementary Table 3).
Group 6 was assigned to specimens with a bottlebrush colony form and a sclerite complement of only scales in the polyp body and scales and sometimes small rods in the tentacle region (Table 1 and Supplementary Table 2). Polyp body scales tend to be figure-8-shaped and there is generally a median constriction regardless of shape. Scales in the tentacle rachis are always transversely arranged in a single series and are often butterfly or bone shaped. In some species, there are pairs of small rods, at the base of the tentacle, arranged either side of the scale. Having scales and occasionally rods places species assigned to Group 6 in Versluys’ sclerite group B or C. This morphological group includes the nominal species C. rigida (Versluys, 1902), C. artospira (Pante and Watling, 2012), C. campanula (Madsen, 1944) and C. tricaulis (Pante and Watling, 2012) (Supplementary Table 3).
Assignment of Group 7 was based on the taxonomic examination of C. cavea (Kinoshita, 1913) and C. geniculata (Wright and Studer, 1889). Specimens assigned to Group 7 all had a bottlebrush colony form with a regular 1/3L branching sequence. The diagnostic character for this group is the presence of extremely long pinnule sclerites, that are toothed on the ventral margin. Pinnule sclerites in Chrysogorgia s.l. are usually small scales, occurring singly or in bundles which are embedded within the tissue of the pinnules. However, the characteristic pinnule sclerites of Group 7 are so long that the tentacles are always extended as opposed to being folded in toward the mouth, allowing these sclerites to be easily seen under a stereo-microscope. Polyp body and tentacle sclerites are scales only, and rods and/or spindles are never present, corresponding with Versluys’ sclerite group C (Table 1).
The Chrysogorgia s.s. clade was designated based on the examination of the type species C. desbonni (Duchassaing and Michelotti, 1864). Morphologically, this clade is characterized by variable colony shape, including bi-flabellate forms. The sclerite complement is rods and/or spindles only, placing this group in Versluys’ sclerite group A. Sclerites in the polyp body are transversely arranged and long, curved to conform to the shape of the polyp body, while tentacle sclerites are shorter and longitudinally arranged. This is the only group in which polyp body sclerites are distinctly transversely arranged. The coenenchyme sclerites in this group are also diagnostic, resembling jigsaw pieces, which vary in length, robustness and warty ornamentation on the surface, both within and between species (Table 1 and Supplementary Table 2). Other nominal species in this group include C. thyrsiformis (Deichmann, 1936), C. fewkesii (Verrill, 1883) and C. multiflora (Deichmann, 1936) (Supplementary Table 3).
Specimens were assigned to Dasygorgia s.s. based on the taxonomic priority and characteristics of the type species D. agassizii (Verrill, 1883). This morphological group is characterized by bottlebrush colonies with variable branching sequence. Polyp body sclerites are scales and rods and/or spindles in the basal part of the tentacles, corresponding with the Versluys’ sclerite group B. Other Chrysogorgia s.l. species that fall into this group include C. comans (Kinoshita, 1913) and C. orientalis (Versluys, 1902).
Molecular Results
Read and Assembly Statistics
The total number of raw reads ranged from 632,849 to 13,296,848 per sample (mean: 5,554,471 ± 2,538,201 SD) (Supplementary Table 4). Removing adapter contamination and low-quality bases using Illumiprocessor reduced the number of reads per sample by 3.33–23.28%, resulting in a mean of 5,273,837 ± 2,431,309 SD trimmed reads per sample. Trimmed reads were assembled into 6,026 to 129,463 contigs per sample (mean: 45,076.81 ± 21,731.42 SD) with a mean length of 416.93 ± 119.6 bp (range: 159.20–685.20) using SPAdes (Supplementary Table 4). Contig coverage ranged from 0.2–48.2% per sample. The highest percentage of bases removed from Illumiprocessor (23.28%) and lowest contig coverage after SPAdes assembly (0.2%), were obtained for the same sample, N038, collected 20 years ago and with low quantity (5.43 ng/μL) and quality (330 bp mean fragment length) DNA at extraction. The oldest sample, S080, collected in 1959 had 12.75% of bases removed and low coverage (1.9%) following SPAdes assembly (Supplementary Table 4).
Locus Recovery
A total of 1,333 UCE loci (1,340 targeted) and 1,682 exon loci (1,700 targeted) were recovered from assembled contigs. Mean length of UCE and exon contigs were 1,063 ± 438 bp (range: 280–1,854 bp) and 1,041 ± 431 bp (range: 289–1,800 bp), respectively. There was a significant difference (Wilcoxon signed rank test: W = 5899, p < 0.001) between the proportion of targeted UCE and exon loci recovered from samples (Supplementary Figure 1). A significantly higher proportion (Wilcoxon signed rank test with constraints: V = 4647, p < 0.001) of UCE loci was recovered per sample but locus recovery was variable with mean values of 766 ± 236 UCEs (range: 41–1,026 loci) and 892 ± 279 exons (range: 40–1,208 loci). UCE and exon locus recovery was significantly positively correlated with DNA extraction concentration (p < 0.001, df = 94) and quality (p < 0.01, df = 94), expressed as mean fragment length (bp) (Figure 2). Locus recovery was also significantly negatively correlated (p < 0.05, df = 94) with time since sample collection (Figure 2). We recovered the lowest numbers of UCE and exon loci from samples 50 and 60 years old, despite relatively good quality and concentration of extracted DNA for the latter sample (S080: 192 bp, 32.15 ng/μL). The 50-year-old sample (N025) had very low quality and concentration (<100 bp, 0.08 ng/μL) DNA at extraction. 72 of the samples (75%) in our set were degraded, with an average fragment length of <300 bp, despite some having high DNA concentrations (165–719 ng/μL) at extraction. Of these 72, 27 samples (28%) were collected within the last 10 years. For the 24 samples (25%) which had high quality (330–17,312 bp) and concentration (mean: 196 ng/μL) DNA at extraction, 11 samples (46%) were collected within the last 5 years. The highest number of loci were recovered from a sample (S139) collected 14 years ago with high quality and concentration of DNA at extraction (3,440 bp, 94.26 ng/μL) (Figure 2 and Supplementary Tables 1, 4).
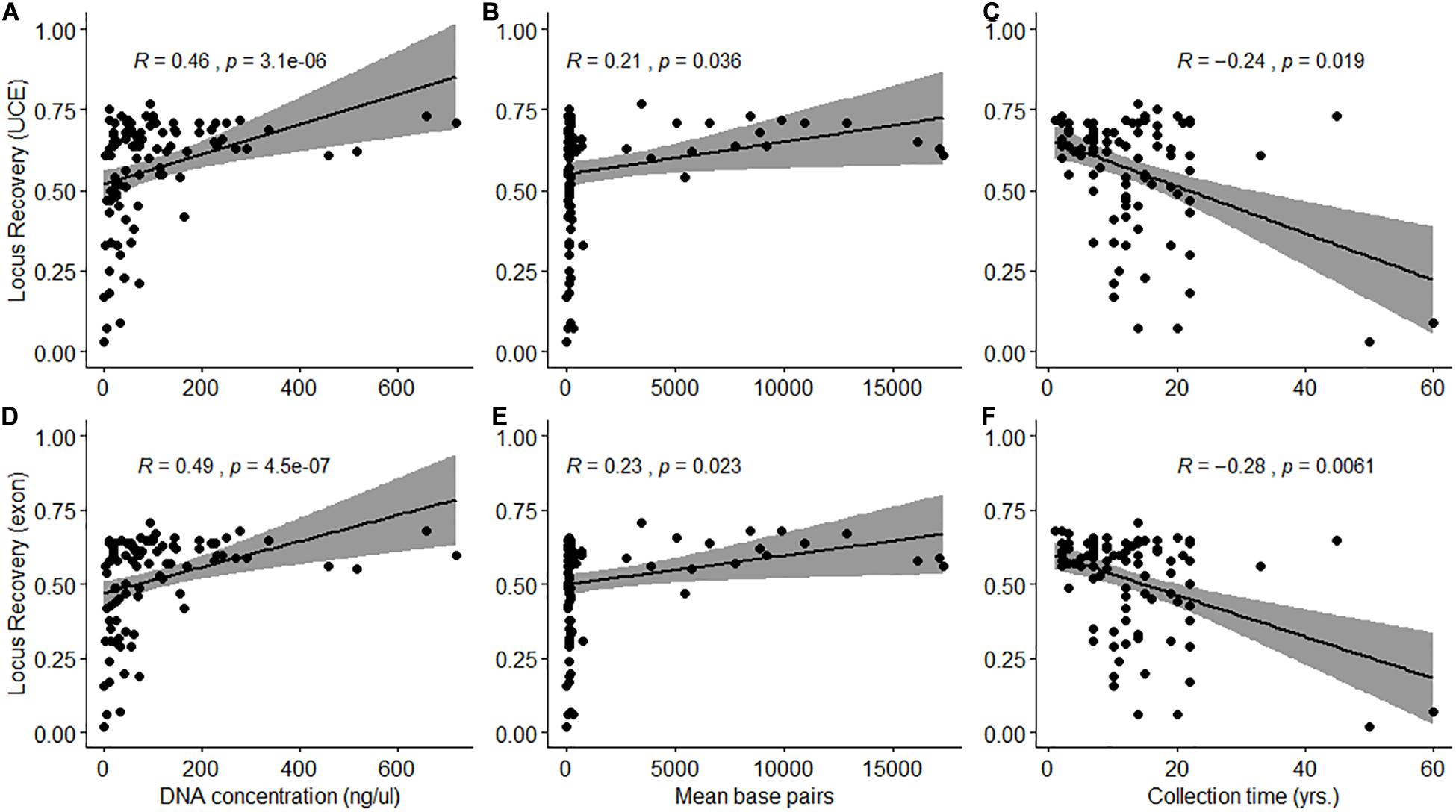
Figure 2. Spearman’s rank correlation between the proportion of loci recovered (A–C: UCE; D–F: Exon) and DNA extraction quantity (ng/μL) and quality (mean base pairs) and time since sample collection (years) (A,D: p < 0.001; B,C,E,F: p < 0.05).
Coverage averaged 0.4 to 278.4x, and 0.4 to 236.3x per sample for the UCE and exon loci set, respectively. There was no significant difference between the median coverage of UCE (94.81x ± 66.07 SD) and exon (83.07x ± 58.24 SD) loci recovered from samples (Wilcoxon signed rank test: W = 4983.5, p = 0.2145). Four samples had loci coverage of <2x for both probe sets. Three of these samples (N025, N038, and S080) also had the lowest recovery (<120) of loci. N025 and N038 were characterized by very low quantity (<5.5 ng/μL), fragmented (<330 bp) DNA, while S80 was collected 60 years ago.
Alignment Statistics
We generated alignment matrices consisting of UCE and exon loci at 50% and 75% occupancy as well as concatenated matrices at each of these occupancies. The alignment matrix generated at 50% with UCE loci had a total of 1,000 loci with a trimmed mean locus length of 686 bp and a total alignment length of 686,391 bp. The alignment matrix generated with the exon loci at 50% occupancy included 1,117 loci, with a trimmed mean locus length of 704 bp and a total length of 786,239 bp. The alignment matrix generated at 75% with UCE loci had a total of 389 loci with a trimmed mean locus length of 731 bp and a total alignment length of 284,292 bp. The alignment matrix generated with the exon loci at 75% occupancy included 336 loci, with a trimmed mean locus length of 733 bp and a total length of 246,168 bp. The concatenated alignment matrix with both loci at 50% occupancy had 2,117 loci with a mean locus length of 695 bp and a total length of 1,470,231 bp. The concatenated alignment matrix with both loci at 75% occupancy had 725 loci with a trimmed mean locus length of 732 bp and a total alignment length of 530,460 bp. The average percentage of parsimony informative sites across all alignments was 35.43%.
Phylogenetic Analysis
Maximum Likelihood topologies produced using the 50 and 75% occupancy datasets were congruent overall (Figure 3). Bootstrap support values were higher overall in the 50% ML tree but in both topologies bootstrap values were 100% for the majority of nodes (Figure 3). Analyses recovered 11 well-supported clades of Chrysogorgia s.l., most of which were congruent with the morphological groups assigned herein. Group 5 was the exception, with samples being divided among three different clades. The clade representing morphological Group 1 was sister to all other morphological groups, which together formed a second well-supported clade. Within the latter clade, Chrysogorgia s.s. and Group 3 are sisters to one another and diverge earliest from a third well-supported clade. A group of specimens assigned to Group 5 are the earliest divergence in this clade and sister to a fourth well-supported clade. Within this clade, a group of specimens assigned to morphological Group 5 diverges first and is sister to a fifth well supported clade. Group 7 is sister to Group 2 and Dasygorgia s.s and diverged before Group 4 and Group 5 and 6, which together represent the most recent lineage.
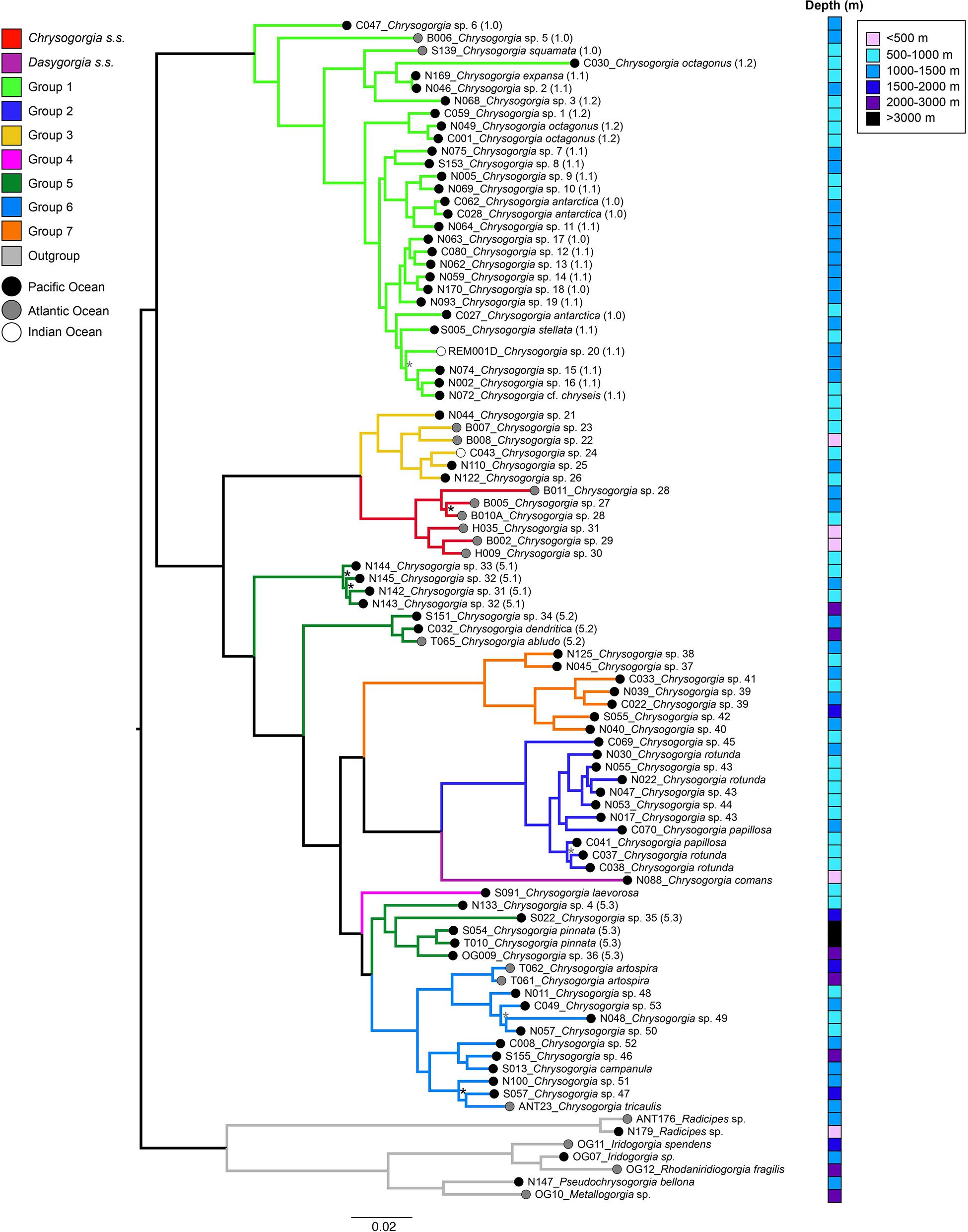
Figure 3. Maximum-likelihood phylogeny constructed with a concatenated UCE and exon loci dataset for 92 taxa at 50% taxon occupancy and rooted to outgroup genera. Branch scale represents expected substitutions per site. Bootstrap support values are <100% for nodes indicated with a * (gray symbol indicates BS < 100% for both 50% and 75%, while black symbol indicates BS < 100% for 75% only). Branches are color coded by morphological group. For Group 1 and Group 5 specimens sub-group assignment is indicated by numbers in brackets. Tip labels are color coded to ocean basin and the depth distribution of each sample is indicated: <500 m; 500–1000 m; 1000–1500 m; 1500–2000 m; 2000–3000 m, and >3000 m.
While specimens were initially assigned to morphological Group 5 based on a broad set of characters, subsequent, detailed morphological examination showed that there are morphological differences which could account for the observed phylogenetic patterns (Table 1 and Figure 3). Clade 5.1 contains a group of specimens, collected from the Pacific, characterized by a pinnate, planar colony form. Polyp body sclerites in this group of specimens are always small, figure 8-shaped scales, while the tentacles can have both rods and scales. The closely related Clade 5.2 contains a type specimen of C. abludo, a species known to have variable colony form. Clade 5.2 also contains a specimen of C. dendritica, with an arborescent colony having a nearly straight main axis and specimen S151 is a planar, pinnate colony. Polyp body sclerites differ from those in Clade 5.1 specimens, in that the polyp body scales are of a highly irregular, ameboid shape and tentacle sclerites are only needle-shaped spindles. Clade 5.3 contains type material of the deep-water species C. pinnata, a planar, pinnate colony, and two other pinnate specimens, that have only rods/spindle-shaped sclerites in the polyp body wall and tentacles (Figure 3 and Supplementary Table 2).
Biogeographic Patterns
Representatives of Group 1 and Group 3 were collected from the Atlantic, Pacific and Indian Oceans. Group 7, Group 2, and two (5.1, 5.3) of the three clades of Group 5 were sampled exclusively from the Pacific, while Chrysogorgia s.s. is an exclusively Atlantic genus (Figure 3). All samples in these clades, were assigned to the same Versluys sclerite group and the sclerite complement was either rods and/or spindles only (A: “Spiculosae”) or scales only (C: “Squamosae”). Diagnostic features related to the colony form and sclerites were also established for these groups. In contrast, groups with representatives from more than one ocean basin tended to have variable colony form, branching sequence and a mix of sclerite types (B: Squamosae abberantes”). Samples from most groups were collected from a wide depth range (500–2000 m). Chrysogorgia s.s., Dasygorgia s.s. and Group 3 were the only groups containing samples collected from depths shallower than 500 m. Only Group 5 (5.2, 5.3) and Group 6 specimens were collected from depths exceeding 2000 m (Figure 3).
Recovery of mtMutS and 28S rDNA
We recovered mtMutS sequences, ranging from partial sequences as short as 87 bp to the entire gene region (2,961 bp), from 50 individuals. We also recovered 96 rDNA sequences ranging from partial 28S sequences as short as 141 bp to sequences as long as 2,890 bp that include not only 28S, but also portions of 18S and/or 5.8S. From the SPAdes assemblies, we also recovered whole mitochondrial genomes for nine specimens and long regions (>8k bp) for five others.
Discussion
We have demonstrated that UCE and exon loci successfully delimit a priori assigned morphological groups within Chrysogorgia s.l., in most cases, and that some of these groups are also well differentiated by depth and ocean basin. We have also demonstrated that targeted enrichment of UCE and exon loci can be successfully applied to older, museum and fluid-preserved specimens of octocorals, although locus recovery is highly variable and significantly related to specimen age, DNA concentration and average fragment length of the extracted DNA fragments.
Implications for Chrysogorgia Taxonomy
Specimens of Chrysogorgia s.l. were assigned to morphological groups prior to sequencing based on characters, purported to be taxonomically important and others which were identified through extensive examination of specimens assigned to species in the genus, including type material. The characters presented here are intentionally broad and do not detail the entirety of fine-scale taxonomic detail used for species designation. Fine-scale taxonomic characters used in the identification of Chrysogorgia s.l. are complex and require expertise and technical jargon to understand. A complete and thorough taxonomic revision is needed to clearly define and document the full complement and range of characters which delineate each group and importantly, Chrysogorgia s.s.
In most instances, there was concordance between assigned morphological groups and highly supported clades recovered from the UCE + exon phylogeny. The exception was specimens from Group 5, initially assigned to one group based on their unusual features compared with all other Chrysogorgia s.l., but which actually fall into three monophyletic clades in our phylogeny. Taxonomic examination of Group 5 specimens a posteriori clearly identified morphological features in colony shape and sclerite complement of polyps and tentacles that define each of these three clades. This highlights the value of an integrated approach using phylogenetic analysis to guide morphological taxonomy and reveal the homoplasious nature of some characters. Similarly, a thorough taxonomic investigation resulted in amended identifications of several samples, some of which have been published under their previous species identifications. DNA barcoding makes use of genetic variation in short, standardized gene regions to aid species identification (Hebert et al., 2003). Many of these studies make use of sequences deposited to online repositories (e.g., BOLD, GenBank) which are linked to vouchered specimens assigned a taxonomic identification. If the morphological identification is incorrect or out of date, the error is propagated, as it is associated with a barcode that is then used to identify additional specimens (Hubert and Hanner, 2015).
Overall, we recognize 11 molecular clades that are also defined by specimen morphology; two of these clades have precedents as Chrysogorgia s.s. and Dasygorgia s.s. The most complete taxonomic treatment of Chrysogorgia s.l. is the monograph published by Versluys (1902), in which he defines branching sequence and suggests, based on the assertions of Wright and Studer (1889), that Chrysogorgia s.l. could be split into three groups on the basis of sclerite types in the polyps and tentacles. He defines three groups: the “Spiculosae” (also called group A), a grouping of species having rods or spindle shaped sclerites, while the “Squamosae” (group C) have scale sclerites in these regions. An intermediate group, the “Squamosae abberantes” (group B) are defined by having scale sclerites in the polyp body but rods in the tentacles. A fourth group, D, was established for two species, C. upsilonia and C. tuberculata, which also have a mix of sclerite types, as in group B, but have rods in the polyp body and scales in the tentacles (Cordeiro et al., 2015). This group has not been formally defined and is considered herein as being part of Versluys’ group B, due to the mix of sclerite types. Branching sequence and sclerite group (A, B, C) are considered useful for distinguishing groups and species in Chrysogorgia s.l., however, analyses reveal that they do not reflect phylogenetic relationships. We found that branching sequence, as defined by Versluys (1902), can be variable within a single colony and often cannot be determined for planar or flabellate colonies, or for fragments of whole bottlebrush colonies, resulting in this character having limited taxonomic utility in Chrysogorgia s.l. Six clades included specimens belonging to more than one Versluys’ sclerite group, while for Group 2, Group 4, Group 7, Chrysogorgia s.s. and Dasygorgia s.s. the same Versluys’ sclerite group could be assigned to all specimens. There were also a few discrepancies in the taxonomic literature with regards to sclerite groups assigned to nominal species and in several cases, we found that our assessment of sclerite groups differed from that assigned previously. For example, C. dendritica is reported as belonging to Versluys’ group A, but the polyp body contains scales, placing this species in sclerite group B (Xu et al., 2020). Examination of type specimens of C. spiculosa revealed the presence of scales in the polyp body, classifying this species in group B, while in the literature it has only been reported as a group A species (Versluys, 1902; Cairns, 2001). For clades where Versluys’ sclerite group was consistent among all specimens, there were important additional sclerite characters diagnostic for the clade. In both Chrysogorgia s.s. and Group 2, sclerites are Versluys’ group A (“Spiculosae”), being rods and/or spindles only. In Chrysogorgia s.s. sclerites can be both rods with rounded ends, and spindles, with acute ends, and in the polyp body sclerites are long and curved, sometimes robust and bearing lateral protuberances. In Group 2, sclerites are always small rods of a standard size and shape, often sparsely distributed in the polyp body. There is scope and need to revise these sclerite groups and update their definitions to include these nuanced characteristics for incorporation into the taxonomic classification of new genera.
Our recognition of 11 morphologically defined groups corresponding with clades recovered in the UCE + exon phylogeny, considered to be putative genera, and 53 forms which could not be matched to existing species, clearly demonstrates the diversity within Chrysogorgia s.l. and highlights the need to consider several complex character states in the revision of this genus. It is clear that, not only the sclerite complement, but also their arrangement, shape and distribution in the polyp body wall, tentacle and pinnules are important characters distinguishing lineages. Morphological characters defining groups are complex, should be considered in combination, and some characters are variable within groups as well as being shared between groups (i.e., colony form and branching sequence). Different families and genera of closely related octocorals are known to share characters such as sclerite form and colony morphology indicating that these characters can be homoplasious and are not necessarily taxonomically important (Figueroa and Baco, 2014; Vargas et al., 2014). This further highlights the inherent difficulty in the taxonomy of octocorals and the need to develop integrated morphological and molecular approaches in revisions and species designations.
There are several instances where specimens assigned to the same nominal or putatively new species do not cluster together, for example specimens C030, C062 and C028 in Group 1 identified as C. antarctica. From a molecular perspective this could be due to introgressive hybridization or incomplete lineage sorting (Quattrini et al., 2019). It is also possible that the current morphological characters used are not diagnostic enough to tease apart cryptic species. In other cases where several morphologically identified species group together (i.e., C. rotunda, C. papillosa, Chrysogorgia sp. 43), it is possible that the morphological characters used are ‘over-splitting’ taxa, or perhaps these characters are phenotypically plastic. A follow-up study using species delimitation methods and quantitative methods for defining morphological differences is necessary to address these outstanding issues.
In a family level investigation, Pante et al. (2012) included 14 specimens of Chrysogorgia s.l. and found this group to be monophyletic based on a concatenated mtMutS, cox1 and 18S region. Our sample set included five (C. chryseis, C. abludo, C. pinnata, C. artospira and C. tricaulis) of the 14 Chrysogorgia analyzed in the mtMutS, cox1 and 18S region phylogenetic study of Pante et al. (2012), three of which were the same specimen. While both studies recovered Chrysogorgia s.l. as monophyletic, our a priori assigned morphological groups, recovered as clades in our UCE + exon phylogeny were individually monophyletic, in most cases. The most comprehensive phylogeny for Chrysogorgia prior to our work is based on RAD-seq data from 82 samples (Pante et al., 2015), assigned to one of 12 mitochondrial haplotypes corresponding with discrete morphologies. However it is difficult to compare their tree against ours because theirs lacks species names or descriptions of morphology (Pante et al., 2015). While mtMutS and 28S regions were not explicitly targeted in our study, we did recover some sequence data for most of our specimens. The ability to coincidentally extract more commonly sequenced regions during target enrichment of UCEs and exons has the added advantage of enabling greater integration between studies using Next Generation and Sanger sequencing.
Biogeography
We identified three groups that corresponded to single ocean distributions. Chrysogorgia s.s. is an exclusively Atlantic genus while Group 2, Group 5.1, 5.3 and Group 7 are exclusively Pacific clades. Representatives of all other Groups were found in both the Atlantic and Pacific Oceans, while only Group 1 and 3 contained Indian Ocean samples as well. A connection between Atlantic and Pacific Ocean Chrysogorgia s.l. based on the direction of branching, Versluys’ sclerite group and mtMutS haplotypes has been reported and hypothesized to have been sustained for much of the evolution of Chrysogorgia s.l., based on species with all major morphological characters being found in both oceans (Pante and Watling, 2012). Region-specific lineages have been found in other octocoral groups. Ellisellid and Melithaeid clades were found to correspond more to geographic distribution than to colony morphology (Bilewitch et al., 2014; Reijnen et al., 2014). Most groups in the present study were collected from a wide depth range (500–2000 m), indicating that they have also successfully diversified across a broad bathymetric gradient. There were, however, a few groups that displayed differentiation in depth range. Chrysogorgia s.s., Dasygorgia s.s. and Group 3 were the only groups which had specimens collected from shallower than 500 m and only Group 5 and Group 6 specimens were collected from depths exceeding 2000 m. We acknowledge that these observations are based on relatively few samples, which could bias results. Further exploration and sampling will more clearly elucidate patterns with depth and geography. Nevertheless, depth and the environmental factors that co-vary with depth have a significant influence on the evolution and ecology of octocorals (e.g., Pante and Watling, 2012; Quattrini et al., 2013; Pante et al., 2015). Pante and Watling (2012) investigated Chrysogorgia s.l. species from the Atlantic and contended that the depth distribution of NW Atlantic species is influenced by the nature of the substrate (hard vs. soft) which can be inferred from the shape of the holdfast. Following formal definition and description of the 11 candidate genera established herein, future work should investigate the evolution and potential habitat association of their morphology.
Utility of This Methodology
We successfully sequenced a high number (99%) of targeted UCE and exon loci overall using an octocoral specific probe set (Erickson et al., 2020). This is the first time that target enrichment of UCE and exon loci has been applied to older (collected > 30 years ago), fluid-preserved museum specimens of octocorals. Here we extracted usable data from specimens that were collected up to 45 years ago and even samples with comparatively poor locus recovery still yielded a large amount of data (e.g., N025: 50 years since collection; 41 and 40 loci for UCE and exons, respectively). Target enrichment of UCE and exon loci has been used in various other invertebrate and vertebrate taxa held in museum collections, for a variety of specimen ages, and histories of preservation and collection (McCormack et al., 2016; Ruane and Austin, 2017; Wood et al., 2018; Tsai et al., 2019). A major advantage of this method is that it can accommodate short, fragmented DNA, inherent in museum samples, that is otherwise a significant impediment for successful Sanger sequencing (Derkarabetian et al., 2019; Tsai et al., 2019). Our findings are significant for phylogenetic studies within the Octocorallia, because the ability to sequence older, low quality type and museum material, particularly for octocorals which have little tissue, means that specimens previously unsuitable for genetic studies in this group can now be examined. The difficulty of sampling fresh octocoral specimens also necessitates reliance on existing collections. Obtaining genetic information from specimens in natural history collections and on which species descriptions have been based is fundamental to documenting and understanding biodiversity and evolutionary processes (Yeates et al., 2016).
While our approach was highly successful, locus recovery was still significantly negatively correlated with time since collection of the specimen, DNA concentration and fragment length. Declines in the quality and quantity of DNA over time are mediated by enzymatic and biochemical processes (Derkarabetian et al., 2019). Notwithstanding our statistical results we found locus recovery to be highly variable. For example, we recovered the lowest numbers of UCE and exon loci from a sample collected 50 years ago and having a very low quality and quantity of DNA at extraction, yet a sample collected 10 years later (i.e., 60 years ago) had over double the number of loci recovered and the concentration of DNA at extraction was 30x higher than the 50 year old sample and more than several other samples collected as recently as 7 years ago. Other factors such as collection practices, preservation history and storage must play a part in the variability of these trends (McCormack et al., 2016). In the past, collection and preservation protocols for octocorals have sometimes employed an initial fixation step using formalin, after which samples are transferred to ethanol for long-term storage (Alderslade and Fabricius, 2001; Etnoyer et al., 2006; Williams and Van Syoc, 2007). Fixation with formalin is well documented to negatively affect both the quality and quantity of DNA which can be obtained from biological specimens (Sarot et al., 2017). Factors such as the duration of exposure to the fixative and environmental conditions such as temperature as well as the duration of the sampling event and time spent on deck will contribute to the degradation of samples and the challenges around extracting PCR-amplifiable DNA from octocorals. All the samples used in this study had been stored in ethanol, but the fixation history and other collection and curation idiosyncrasies are unknown and generally not documented in museum data, which inhibits our ability to understand their effects on DNA analysis.
Conclusion
Our phylogenetic results form the basis to propose that 11 candidate genera, with specific morphologies, should be recognized. The multi-locus approach used in this study has significantly improved our understanding of diversity and will guide the revision of this morphologically complex group. Fully resolving the taxonomy of the Chrysogorgiidae will require a comprehensive assessment of nominal species and candidate genera. Because this study sequenced just a fraction of the species of Chrysogorgiidae, more individuals and more putative species will help elucidate species-level diversity within this family. To help delimit species, combining coalescent-based and allele frequency methods with SNP calling from the UCE and exon loci will be considered for future work. Target locus enrichment for octocorals will revolutionize systematics studies in this group and facilitate resolution of deep phylogenetic relationships which is not possible using mitochondrial regions and Sanger sequencing. We also demonstrated that targeted bait enrichment of UCEs and exons is suitable for obtaining sequence data from older, museum and fluid preserved specimens.
Data Availability Statement
The datasets generated in this study can be found in NCBI under submission number SUB8651496 and the BioProject ID PRJNA681648, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA681648.
Author Contributions
CU acquired the data, performed the taxonomic analyses, and led the writing. CU, AQ, and CM performed the molecular analyses. PA trained CU in octocoral taxonomy. All authors made substantial contributions to discussing the methods, results and revising the manuscript, and designed the study.
Funding
This work originates from a project that is funded, collectively, by the University of Tasmania and CSIRO Oceans and Atmosphere. Project data were collected during several research trips to institutes funded through grants provided by the Smithsonian Institute, Museum of Comparative Zoology, Yale Peabody Museum, NIWA and Harvey Mudd College and during voyages on the Southern Surveyor and Australia’s Marine National Facility Vessel, RV Investigator, funded through the Australian Government. Funding for this work came from several sources: CSIRO Oceans and Atmosphere, University of Tasmania, Smithsonian Institute, Museum of Comparative Zoology, Yale Peabody Museum and NSF-DEB 1457817 to CM. None of these funding agencies played any role in the study design; collection, interpretation or analysis of data; or writing of this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank the managers, curators and collections staff of CSIRO Oceans and Atmosphere, NIWA Invertebrate Collection, Smithsonian Institute, Museum of Comparative Zoology, Yale Peabody Museum, National Museum of Brazil for access to samples held in their institutes for taxonomic and genetic work. Harvey Mudd College provided facilities needed to undertake data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.599984/full#supplementary-material
Footnotes
- ^ http://phyluce.readthedocs.io/en/latest (accessed March 2019).
References
Alderslade, P. A. (1998). Revisionary systematics in the gorgonian family Isididae, with descriptions of numerous new taxa (Coelenterata: Octocorallia). Rec. West. Aust. Mus. 55, 1–359. doi: 10.1007/978-3-319-23534-9_1
Alderslade, P. A., and Fabricius, K. (2001). Soft Corals and Sea Fans. Australian: Australian Institute of Marine Science, 1–264.
Althaus, F., Williams, A., Schlacher, T. A., Kloser, R. J., Green, M. A., Barker, B. A., et al. (2009). Impacts of bottom trawling on deep-coral ecosystems of seamounts are long-lasting. Mar. Ecol. Prog. Ser. 397, 279–294. doi: 10.3354/meps08248
Ament-Velásquez, S. L., Breedy, O., Cortés, J., Guzman, H. M., Wörheide, G., and Vargas, S. (2016). Homoplasious colony morphology and mito-nuclear phylogenetic discordance among Eastern Pacific octocorals. Mol. Phylogenet. Evol. 98, 373–381. doi: 10.1016/j.ympev.2016.02.023
Auster, P. J., Gjerde, K., Heupel, E., Watling, L., Grehan, A., and Rogers, A. D. (2011). Definition and detection of vulnerable marine ecosystems on the high seas: problems with the “move-on” rule. ICES J. Mar. Sci. 68, 254–264. doi: 10.1093/icesjms/fsq074
Baco, A. R., and Cairns, S. D. (2012). Comparing molecular variation to morphological species designations in the deep-sea coral Narella reveals new insights into seamount coral ranges. PLoS One 7:e45555. doi: 10.1371/journal.pone.0045555
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bayer, F. M., and Stefani, J. (1988). A new species of Chrysogorgia (Octocorallia: Gorgonacea) from new Caledonia, with descriptions of some other species from the Western Pacific. Proc. Biol. Soc. Wash. 101, 257–279.
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Frizsch, G., et al. (2013). MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. doi: 10.1016/j.ympev.2012.08.023
Bilewitch, J. P., Ekins, M., Hooper, J., and Degnan, S. M. (2014). Molecular and morphological systematics of the Ellisellidae (Coelenterata: Octocorallia): parallel evolution in a globally distributed family of octocorals. Mol. Phylogenet. Evol. 73, 106–118. doi: 10.1016/j.ympev.2014.01.023
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cairns, S. D. (2001). Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa) part 1: the genus chrysogorgia Duchassaing & Michelotti, 1864. Proc. Biol. Soc. Wash. 114, 746–787.
Cairns, S. D. (2002). “A new species of Chrysogorgia (Anthozoa: Octocorallia) from the Antarctic,” in Proceedings of the Biological Society of Washington, 115, 217–222.
Cairns, S. D. (2018). Deep-water octocorals (Cnidaria, Anthozoa) from the Galápagos and Cocos islands. Part 1: suborder Calcaxonia. Zookeys 2018, 1–46. doi: 10.3897/zookeys.729.21779
Collins, R. A., and Hrbek, T. (2018). An in silico comparison of protocols for dated phylogenomics. Syst. Biol. 67, 633–650. doi: 10.1093/sysbio/syx089
Cordeiro, R. T. S., Castro, C. B., and Peres, C. D. (2015). Deep-water octocorals (Cnidaria: Octocorallia) from Brazil: family Chrysogorgiidae. Zootaxa 4058, 81–100. doi: 10.11646/zootaxa.4058.1.4
De Clippele, L. H., Huvenne, V. A. I., Molodtsova, T. N., and Roberts, J. M. (2019). The diversity and ecological role of non-scleractinian corals (Antipatharia and Alcyonacea) on scleractinian cold-water coral mounds. Front. Mar. Sci. 6:16. doi: 10.3389/fmars.2019.00184
Deichmann, E. (1936). “The Alcyonaria of the Western part of the Atlantic Ocean,” Memoirs of the Museum of Comparative Zoology, Cambridge, United states, Printed for the Museum, 53, 253–308.
Derkarabetian, S., Benavides, L. R., and Giribet, G. (2019). Sequence capture phylogenomics of historical ethanol-preserved museum specimens: unlocking the rest of the vault. Mol. Ecol. Resour. 19, 1531–1544. doi: 10.1111/1755-0998.13072
Duchassaing, P., and Michelotti, J. (1864). Supplément au mémoire sur les coralliaires des Antilles. Memorie della Reale Accademia della Scienze di Torino, Serie 2 23, 1–23. Available online at: https://www.biodiversitylibrary.org/page/11762431
Eaton, D. A. R., Spriggs, E. L., Park, B., and Donoghue, M. J. (2017). Misconceptions on missing data in RAD-seq phylogenetics with a deep-scale example from flowering plants. Syst. Biol. 66, 399–412.
Erickson, K., Pentico, K., Quattrini, A. M., and McFadden, C. S. (2020). New approaches to species delimitation and population structure of anthozoans: two case studies of octocorals using ultraconserved elements and exons. Mol. Ecol. Resour. 21, 78–92. doi: 10.1101/2020.04.01.021071
Etnoyer, P. J., Cairns, S. D., Sanchez, J. A., Reed, J. K., Lopez, J. V., Schroeder, W. W., et al. (2006). Deep-Sea Coral Collection Protocols: a Synthesis of Field Experience from Deep-Sea Coral Researchers, Designed to Build our National Capacity to Document Deep-Sea Coral Diversity. Silver Spring, MD: NOAA National Marine Fisheries Service.
Faircloth, B. C. (2013). Illumiprocessor: A Trimmomatic Wrapper for Parallel Adapter and Quality Trimming. Available online at: http://dx.doi.org/10.6079/J9ILL
Faircloth, B. C. (2016). PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32, 786–788. doi: 10.1093/bioinformatics/btv646
Faircloth, B. C., Branstetter, M. G., White, N. D., and Brady, S. G. (2015). Target enrichment of ultraconserved elements from arthropods provides a genomic perspective on relationships among hymenoptera. Mol. Ecol. Resour. 15, 489–501. doi: 10.1111/1755-0998.12328
Faircloth, B. C., McCormack, J. E., Crawford, N. G., Harvey, M. G., Brumfield, R. T., and Glenn, T. C. (2012). Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 61, 717–726. doi: 10.1093/sysbio/sys004
Faircloth, B. C., Sorenson, L., Santini, F., and Alfaro, M. E. (2013). A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs). PLoS One 8:e65923. doi: 10.1371/journal.pone.0065923
FAO (2009). Report of the Technical Consultation on International Guidelines for the Management of Deep-Sea Fisheries in the High Seas. Rome: FAO Fisheries and Aquaculture Report.
Figueroa, D. F., and Baco, A. R. (2014). Octocoral mitochondrial genomes provide insights into the phylogenetic history of gene order rearrangements, order reversals, and cnidarian phylogenetics. Genome Biol. Evol. 7, 391–409. doi: 10.1093/gbe/evu286
France, S. C., Rosel, P. E., Agenbroad, J. E., Mullineaux, L. S., and Kocher, T. D. (1996). DNA sequence variation of mitochondrial large-subunit rRNA provides support for a two-subclass organization of the Anthozoa (Cnidaria). Mol. Mar. Biol. Biotechnol. 5, 15–28.
Harvey, M. G., Smith, B. T., Glenn, T. C., Faircloth, B. C., and Brumfield, R. T. (2016). Sequence capture versus restriction site associated DNA sequencing for shallow systematics. Syst. Biol. 65, 910–924. doi: 10.1093/sysbio/syw036
Hebert, P. D. N., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. Lond. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hubert, N., and Hanner, R. (2015). DNA barcoding, species delineation and taxonomy: a historical perspective. DNA Barcodes 3, 44–58.
Kinoshita, K. R. (1913). Studien uber einige Chrysogorgiiden Japans. J. Coll. Sci. Tokyo Imp. Univ. 33, 1–47.
Kükenthal, W. (1909). Diagnosen neuer Gorgoniden aus der Gattung Chrysogorgia (6. Mitteilung.). Zool. Anzeiger 33, 704–708.
Leaché, A. D., Chavez, A. S., Jones, L. N., Grummer, J. A., Gottscho, A. D., and Linkem, C. W. (2015). Phylogenomics of phrynosomatid lizards: conflicting signals from sequence capture versus restriction site associated DNA sequencing. Genome Biol. Evol. 7, 706–719. doi: 10.1093/gbe/evv026
McCormack, J. E., Tsai, W. L. E., and Faircloth, B. C. (2016). Sequence capture of ultraconserved elements from bird museum specimens. Mol. Ecol. Resour. 16, 1189–1203. doi: 10.1111/1755-0998.12466
McFadden, C. S., Benayahu, Y., Pante, E., Thoma, J. N., Nevarez, P. A., and France, S. C. (2010). Limitations of mitochondrial gene barcoding in Octocorallia. Mol. Ecol. Resour. 11, 19–31. doi: 10.1111/j.1755-0998.2010.02875.x
McFadden, C. S., Brown, A. S., Brayton, C., Hunt, C. B., and van Ofwegen, L. P. (2014a). Application of DNA barcoding in biodiversity studies of shallow-water octocorals: molecular proxies agree with morphological estimates of species richness in Palau. Coral Reefs 33, 275–286. doi: 10.1007/s00338-013-1123-0
McFadden, C. S., France, S. C., Sanchez, J. A., and Alderslade, P. (2006). A molecular phylogenetic analysis of the Octocorallia (Cnidaria: Anthozoa) based on mitochondrial protein-coding sequences. Mol. Phylogenet. Evol. 41, 513–527. doi: 10.1016/j.ympev.2006.06.010
McFadden, C. S., Haverkort-Yeh, R., Reynolds, A. M., Halàsz, A., Quattrini, A. M., Forsman, Z. H., et al. (2017). Species boundaries in the absence of morphological, ecological or geographical differentiation in the Red Sea octocoral genus Ovabunda (Alcyonacea: Xeniidae). Mol. Phylogenet. Evol. 112, 174–184. doi: 10.1016/j.ympev.2017.04.025
McFadden, C. S., Reynolds, A. M., and Janes, M. P. (2014b). DNA barcoding of xeniid soft corals (Octocorallia: Alcyonacea: Xeniidae) from Indonesia: species richness and phylogenetic relationships. Syst. Biodivers. 12, 247–257. doi: 10.1080/14772000.2014.902866
Mosher, C. V., and Watling, L. (2009). Partners for life: a brittle star and its octocoral host. Mar. Ecol. Prog. Ser. 397, 81–88. doi: 10.3354/meps08113
Pante, E., Abdelkrim, J., Viricel, A., Gey, D., France, S. C., Boisselier, M. C., et al. (2015). Use of RAD sequencing for delimiting species. Heredity 114, 450–459. doi: 10.1038/hdy.2014.105
Pante, E., and France, S. C. (2010). Pseudochrysogorgia bellona n. gen., n. sp.: a new genus and species of chrysogorgiid octocoral (Coelenterata, Anthozoa) from the Coral Sea. Zoosystema 32, 595–612. doi: 10.5252/Z2010N4A4
Pante, E., and Watling, L. (2012). Chrysogorgia from the New England and corner seamounts: Atlantic-Pacific connections. J. Mar. Biol. Assoc. UK 92, 911–927. doi: 10.1017/S0025315411001354
Pante, E., France, S. C., Couloux, A., Cruaud, C., McFadden, C. S., Samadi, S., et al. (2012). Deep-sea origin and in-situ diversification of chrysogorgiid octocorals. PLoS One 7:e38357. doi: 10.1371/journal.pone.0038357
Quattrini, A. M., Faircloth, B. C., Dueñas, L. F., Bridge, T. C. L., Brugler, M. R., Calixto-Botía, I. F., et al. (2018). Universal target-enrichment baits for anthozoan (Cnidaria) phylogenomics: new approaches to long-standing problems. Mol. Ecol. Resour. 18, 281–295. doi: 10.1111/1755-0998.12736
Quattrini, A. M., Georgian, S. M., Byrnes, L., Stevens, A., Falco, R., and Cordes, E. E. (2013). Niche divergence by deep-sea octocorals in the genus Callogorgia across the upper continental slope of the Gulf of Mexico. Mol. Ecol. 22, 4123–4140. doi: 10.1111/mec.12370
Quattrini, A. M., Rodriguez, E., Faircloth, B. C., Cowman, P. F., Brugler, M. R., Farfan, G. A., et al. (2020). Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 4, 1531–1538. doi: 10.1038/s41559-020-01291-1
Quattrini, A. M., Wu, T., Soong, K., Jeng, M. S., Benayahu, Y., and McFadden, C. S. (2019). A next generation approach to species delimitation reveals the role of hybridization in a cryptic species complex of corals. BMC Evol. Biol. 19:116. doi: 10.1186/s12862-019-1427-y
Ramirez-Llodra, E., Brandt, A., Danovaro, R., De Mol, B., Escobar, E., German, C. R., et al. (2010). Deep, diverse and definitely different: unique attributes of the world’s largest ecosystem. Biogeosciences 7, 2851–2899. doi: 10.5194/bg-7-2851-2010
Reijnen, B. T., McFadden, C. S., Hermanlimianto, Y. T., and van Ofwegen, L. P. (2014). A molecular and morphological exploration of the generic boundaries in the family Melithaeidae (Coelenterata: Octocorallia) and its taxonomic consequences. Mol. Phylogenet. Evol. 70, 383–401. doi: 10.1016/j.ympev.2013.09.028
Roberts, J., Wheeler, A., Freiwald, A., and Cairns, S. (2009). Cold-Water Corals: The Biology and Geology of Deep-Sea Coral Habitats, Cambridge: Cambridge University Press. doi: 10.1017/CBO9780511581588
Ruane, S., and Austin, C. C. (2017). Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Mol. Ecol. Resour. 17, 1003–1008. doi: 10.1111/1755-0998.12655
Sarot, E., Carillo-Baraglioli, M.-F., Duranthon, F., Péquignot, A., and Pyronnet, S. (2017). Assessment of alternatives to environmental toxic formalin for DNA conservation in biological specimens. Environ. Sci. Pollut. Res. 24, 16985–16993. doi: 10.1007/s11356-017-9349-y
Schnabel, K. E. (2020). The marine fauna of New Zealand. Squat lobsters (Crustacea, Decapoda, Chirostyloidea). NIWA Biodivers. Mem. 132, 1–351.
Smith, B. T., Harvey, M. G., Faircloth, B. C., Glenn, T. C., and Brumfield, R. T. (2014). Target capture and massively parallel sequencing of ultraconserved elements for comparative studies at shallow evolutionary time scales. Syst. Biol. 63, 83–95. doi: 10.1093/sysbio/syt061
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stearns, R. E. C. (1883). “Descriptions of a new genus and species of alcyonoid polyp, from Japanese waters, with remarks on the structure and habits of related forms, etc.,” in Proceedings of the United States National Museum 6, 96–101. doi: 10.5479/si.00963801.6-346.96
Thoma, J. N., Pante, E., Brugler, M. R., and France, S. C. (2009). Deep-sea octocorals and antipatharians show no evidence of seamount-scale endemism in the NW Atlantic. Mar. Ecol. Prog. Ser. 397, 25–35. doi: 10.3354/meps08318
Thomson, J. A., and Henderson, W. D. (1906). An Account of the Alcyonarians Collected by the Royal Indian Marine Survey Ship ‘Investigator’ in the Indian Ocean. I. The Alcyonarians of the Deep Sea. Trustees of the Indian Museum, University of Aberdeen, 1–128. Available online at: https://www.biodiversitylibrary.org/page/12037643
Tsai, W. L. E., Mota-Vargas, C., Rojas-Soto, O., Bhowmik, R., Liang, E. Y., Maley, J. M., et al. (2019). Museum genomics reveals the speciation history of Dendrortyx wood-partridges in the Mesoamerican highlands. Mol. Phylogenet. Evol. 136, 29–34. doi: 10.1016/j.ympev.2019.03.017
Vargas, S., Guzman, H. M., Breedy, O., and Wörheide, G. (2014). Molecular phylogeny and DNA barcoding of tropical eastern Pacific shallow-water gorgonian octocorals. Mar. Biol. 161, 1027–1038. doi: 10.1007/s00227-014-2396-8
Verrill, A. E. (1883). Report on the Anthozoa and on some additional species dredged by the “Blake” in 1877-1879 and by the U. S. Fish Commission Steamer “Fish Hawk” in 1880-1882. Bull. Mus. Comp. Zool. Harv. 11, 1–72.
Versluys, J. (1902). Die gorgoniden der siboga-expedition. i. die Chrysogorgiiden. Siboga Expeditie. 13, 1–120.
Watling, L. (2007). A review of the genus Iridogorgia (Octocorallia: Chrysogorgiidae) and its relatives, chiefly from the North Atlantic Ocean. J. Mar. Biol. Assoc. UK. 87, 393–402. doi: 10.1017/s002531540705535x
Watling, L., and Auster, P. J. (2017). Seamounts on the high seas should be managed as vulnerable marine ecosystems. Front. Mar. Sci. 4:14. doi: 10.3389/fmars.2017.00014
Watling, L., France, S. C., Pante, E., and Simpson, A. (2011). Biology of deep-water octocorals. Adv. Mar. Biol. 60, 41–122. doi: 10.1016/B978-0-12-385529-9.00002-0
Wicksten, M. K. (2020). One to a customer: associations between squat lobsters (Chirostyloidea) and their anthozoan hosts. Mar. Biodivers. 50:14. doi: 10.1007/s12526-019-01035-w
Williams, G. C., and Van Syoc, R. J. (2007). “Methods of preservation and anesthetization of marine invertebrates,” in The Light and Smith Manual: Intertidal Invertebrates from Central California to Oregon, 4th Edn, ed. J. T. Carlton (Berkeley, CA: University of California Press), 37–41. doi: 10.1525/9780520930438-010
Wood, H. M., González, V. L., Lloyd, M., Coddington, J., and Scharff, N. (2018). Next-generation museum genomics: phylogenetic relationships among palpimanoid spiders using sequence capture techniques (Araneae: Palpimanoidea). Mol. Phylogenet. Evol. 127, 907–918. doi: 10.1016/j.ympev.2018.06.038
Wright, E. P., and Studer, T. (1889). Report on the Alcyonaria Collected by H.M.S. Challenger During the Years 1873–76. Available online at: http://www.19thcenturyscience.org/HMSC/HMSC-Reports/Zool-64/htm/doc.html
Xu, Y., Zahn, Z., and Xu, K. (2020). Morphology and molecular phylogeny of three new deep-sea species of Chrysogorgia (Cnidaria, Octocorallia) from seamounts in the tropical Western Pacific Ocean. Peer J. 8:e8832. doi: 10.7717/peerj.8832
Keywords: target-capture, integrative taxonomy, ultra conserved elements, exons, gorgonian
Citation: Untiedt CB, Quattrini AM, McFadden CS, Alderslade PA, Pante E and Burridge CP (2021) Phylogenetic Relationships Within Chrysogorgia (Alcyonacea: Octocorallia), a Morphologically Diverse Genus of Octocoral, Revealed Using a Target Enrichment Approach. Front. Mar. Sci. 7:599984. doi: 10.3389/fmars.2020.599984
Received: 28 August 2020; Accepted: 09 December 2020;
Published: 12 January 2021.
Edited by:
Chiara Romano, Center for Advanced Studies of Blanes (CSIC), SpainReviewed by:
Ana Riesgo, Natural History Museum, United KingdomEkin Tilic, University of Bonn, Germany
Federica Costantini, University of Bologna, Italy
Copyright © 2021 Untiedt, Quattrini, McFadden, Alderslade, Pante and Burridge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Candice Bobby Untiedt, candice.untiedt@csiro.au
 Candice Bobby Untiedt
Candice Bobby Untiedt Andrea M. Quattrini
Andrea M. Quattrini Catherine S. McFadden
Catherine S. McFadden Phil A. Alderslade
Phil A. Alderslade Eric Pante
Eric Pante Christopher P. Burridge
Christopher P. Burridge