Abstract
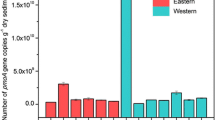
Anammox bacteria have been detected in various ecosystems, but their occurrence and community composition along the shipping channels have not been reported. In this study, anammox bacteria were recovered by PCR-amplified biomarker hzsB gene from the genomic DNA of the sediment samples. Phylogenetic tree revealed that Candidatus Scalindua and Ca. Brocadia dominated the anammox community of the Hong Kong channels; Ca. Scalindua spp. was present abundantly at the sites farther from the shore, whereas Ca. Jettenia and Ca. Kuenenia were detected as the minor members in the estuarine sediments near the shipping terminals. The highest values of Shannon–Wiener index and Chao1 were identified in the sediments along the Urmston road (UR), suggesting the highest α-diversity and species richness of anammox bacteria. PCoA analysis indicated that anammox bacterial communities along UR and Tai Hong (TH) channel were site-specific because these samples were grouped and clearly separated from the other samples. The maximum diversity of anammox bacteria was detected in UR samples, ranging from 6.28 × 105 to 1.28 × 106 gene copies per gram of dry sediment. A similar pattern of their transcriptional activities was also observed among these channels. Pearson’s moment correlation and redundancy analysis indicated that NH4+-N was a strong factor shaping the community structure, which showed significant positive correlation with the anammox bacterial abundance and anammox transcriptional activities (p < 0.01, r > 0.8). Also, NH4+-N, (NO3− + NO2−)-N, and NH4+/NOX were additional key environmental factors that influenced the anammox community diversity and distribution. This study yields a better understanding of the ecological distribution of anammox bacteria and the dominant genera in selective niche.






Similar content being viewed by others
Data availability
Nucleotide sequence data of this study are deposited in NCBI public available database for full access.
References
Amano T, Yoshinaga I, Okada K, Yamagishi T, Ueda S, Obuchi A, Sako Y, Suwa Y (2007) Detection of anammox activity and diversity of anammox bacteria-related 16S rRNA genes in coastal marine sediment in Japan. Microbes Environ 22:232–242. https://doi.org/10.1264/jsme2.22.232
Azari M, Walter U, Rekers V, Gu JD, Denecke M (2017) More than a decade of experience of landfill leachate treatment with a full-scale anammox plant combining activated sludge and activated carbon biofilm. Chemosphere 174:117–126
Bessonet, M, Tiruta-Barna, L, Spérandio, M, 2017. Environmental assessment of anammox process in mainstream with WWTP modeling coupled to life cycle assessment, in: Frontiers in wastewater treatment and modelling, lecture notes in civil engineering. Springer International Publishing, Cham, pp. 392–397. https://doi.org/10.1007/978-3-319-58421-8_62
Census and Statistics Department Hong Kong, 2016. The Profile of Hong Kong Population Analysed by District Council District, 2015 1–9.
Chan, AA, 2015. Experience with Anammox Sludge Liquor Treatment and Up-Coming Challenges with Thermal Hydrolysis Effluents at the Sundet Wwtp, VÄXjÖ Sweden. Proceedings of the Water Environment Federation 2015, 1–11. https://doi.org/10.2175/193864715819558181
Chan HW, Meng H, Gu J-D (2016) Anammox bacteria detected in fish intestinal tract systems. Appl Environ Biotechnol 1(1):13–18
Chao, A, 1984. Nonparametric estimation of the number of classes in a population. Scand J Stat. https://doi.org/10.2307/4615964
Chen J, Gu J-D (2017) Faunal burrows alter the diversity, abundance, and structure of AOA, AOB, anammox and n-Damo communities in coastal mangrove sediments. Microbial Eology 74(1):140–156
Chen L, Liu F, Jia F, Hu Y-J, Lai C, Li X, Luo P, Xiao R-L, Li Y, Wu J-S (2017) Anaerobic ammonium oxidation in sediments of surface flow constructed wetlands treating swine wastewater. Appl Microbiol Biotechnol 101:1301–1311. https://doi.org/10.1007/s00253-016-7957-y
Dalsgaard T, Thamdrup B, Canfield DE (2005) Anaerobic ammonium oxidation (anammox) in the marine environment. Res Microbiol 156:457–464. https://doi.org/10.1016/j.resmic.2005.01.011
Dang H, Li J, Chen R, Wang L, Guo L, Zhang Z, Klotz MG (2010) Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl Environ Microbiol 76:4691–4702. https://doi.org/10.1128/AEM.02563-09
Dang H, Zhou H, Zhang Z, Yu Z, Hua E, Liu X, Jiao N (2013) Molecular detection of Candidatus Scalindua pacifica and environmental responses of sediment anammox bacterial community in the Bohai Sea, China. PLoS One 8:e61330. https://doi.org/10.1371/journal.pone.0061330
Diaz RJ, Rosenberg R (2008) Spreading dead zones and consequences for marine ecosystems. Science 321:926–929. https://doi.org/10.1126/science.1156401
Engström P, Dalsgaard T, Hulth S, Aller RC (2005) Anaerobic ammonium oxidation by nitrite (anammox): Implications for N2 production in coastal marine sediments. Geochimica et Cosmochimica Acta 69:2057–2065. https://doi.org/10.1016/j.gca.2004.09.032
Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA (2008) Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320:889–892. https://doi.org/10.1126/science.1136674
Gruber N, Galloway JN (2008) An earth-system perspective of the global nitrogen cycle. Nature 451:293–296. https://doi.org/10.1038/nature06592
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Sympos Ser 41:95–98
Hammer, Ø, Harper, D, Ryan, PD, 2001. PAST-palaeontological statistics, ver. 1.89. Palaeontologia Electronica
Han P, Gu J-D (2013) More refined diversity of anammox bacteria recovered and distribution in different ecosystems. Appl Microbiol Biotechnol 97:3653–3663. https://doi.org/10.1007/s00253-013-4756-6
Han P, Gu J-D (2015) Further analysis of anammox bacterial community structures along an anthropogenic nitrogen-input gradient from the riparian sediments of the pearl river delta to the deep-ocean sediments of the South China Sea. Geomicrobiol J 32:789–798. https://doi.org/10.1080/01490451.2014.1001502
Han P, Klümper U, Wong A, Li M, Lin J-G, Quan Z, Denecke M, Gu J-D (2017) Assessment of molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in different environmental samples using PCR primers based on 16S rRNA and functional genes. Appl Microbiol Biotechnol 101:7689–7702. https://doi.org/10.1007/s00253-017-8502-3
Harhangi HR, Le Roy M, van Alen T, Hu B-L, Groen J, Kartal B, Tringe SG, Quan Z-X, Jetten MSM, Op Den Camp HJM (2012) Hydrazine synthase, a unique phylomarker with which to study the presence and biodiversity of anammox bacteria. Appl Environ Microbiol 78:752–758. https://doi.org/10.1128/AEM.07113-11
Hou L, Zheng Y, Liu M, Gong J, Zhang X, Yin G, You L (2013) Anaerobic ammonium oxidation (anammox) bacterial diversity, abundance, and activity in marsh sediments of the Yangtze Estuary. J Geophys Res: Biogeosci 118:1237–1246. https://doi.org/10.1002/jgrg.20108
Hou L, Zheng Y, Liu M, Li X, Lin X, Yin G, Gao J, Deng F, Chen F, Jiang X (2015) Anaerobic ammonium oxidation and its contribution to nitrogen removal in China’s coastal wetlands. Nat Publ Group 5:15621. https://doi.org/10.1038/srep15621
Hu B, Shen L, Ping DU, Zheng P, Xu X, Zeng J (2012a) The influence of intense chemical pollution on the community composition, diversity and abundance of anammox bacteria in the Jiaojiang Estuary (China). PLOS ONE 7:e33826. https://doi.org/10.1371/journal.pone.0033826
Hu B-L, Shen L-D, Zheng P, Hu A-H, Chen T-T, Cai C, Liu S, Lou L-P (2012b) Distribution and diversity of anaerobic ammonium-oxidizing bacteria in the sediments of the Qiantang River. Environ Microbiol Rep 4:540–547. https://doi.org/10.1111/j.1758-2229.2012.00360.x
Humbert S, Tarnawski S, Fromin N, Mallet M-P, Aragno M, Zopfi J (2009) Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. The ISME J 4:450–454. https://doi.org/10.1038/ismej.2009.125
Humbert S, Zopfi J, Tarnawski S-E (2012) Abundance of anammox bacteria in different wetland soils. Environ Microbiol Rep 4:484–490. https://doi.org/10.1111/j.1758-2229.2012.00347.x
Jetten MSM, Niftrik LV, Strous M, Kartal B, Keltjens JT, Op Den Camp HJM (2009) Biochemistry and molecular biology of anammox bacteria. Crit Rev Biochem Mol Biol 44:65–84. https://doi.org/10.1080/10409230902722783
Kartal B, Kuypers MMM, Lavik G, Schalk J, Op Den Camp HJM, Jetten MSM, Strous M (2007) Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ Microbiol 9:635–642. https://doi.org/10.1111/j.1462-2920.2006.01183.x
Kartal B, van Niftrik L, Rattray J, van de Vossenberg JLCM, Schmid MC, Sinninghe Damsté J, Jetten MSM, Strous M (2008) Candidatus “Brocadia fulgida”: an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol Ecol 63:46–55. https://doi.org/10.1111/j.1574-6941.2007.00408.x
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability Mol. Biol. Evol. 30:772–780
Kindaichi T, Tsushima I, Ogasawara Y, Shimokawa M, Ozaki N, Satoh H, Okabe S (2007) In situ activity and spatial organization of anaerobic ammonium-oxidizing (anammox) bacteria in biofilms. Appl. Environ. Microbiol. 73:4931–4939. https://doi.org/10.1128/AEM.00156-07
Kong L, Jing H, Kataoka T, Buchwald C, Liu H (2013) Diversity and spatial distribution of hydrazine oxidoreductase (hzo) gene in the oxygen minimum zone off Costa Rica. PLOS ONE 8:e78275. https://doi.org/10.1371/journal.pone.0078275
Kumar S, Tamura K, Nei M (1994) MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10:189–191
Kwan, K-LX, 1998. Hong Kong international port terminal. 10.5353/th_b3198392
Lai J (2013) Canoco 5: a new version of an ecological multivariate data ordination program. Biodivers Sci 21:765–768. https://doi.org/10.3724/SP.J.1003.2013.04133
Lee SY (2016) From blue to black: anthropogenic forcing of carbon and nitrogen influx to mangrove-lined estuaries in the South China Sea. Mar Pollut Bull 109:682–690. https://doi.org/10.1016/j.marpolbul.2016.01.008
Li M, Gu J-D (2016a) The diversity and distribution of anammox bacteria in the marine aquaculture zones. Appl Microbiol Biotechnol 100:8943–8953. https://doi.org/10.1007/s00253-016-7690-6
Li M, Gu J-D (2016b) Molecular evidence of the existence of anaerobic ammonia oxidation bacteria in the gut of polychaete (Neanthes glandicincta). Appl Environ Biotechnol 1(1):19–29
Li M, Hong Y, Cao H, Gu J-D (2013) Community structures and distribution of anaerobic ammonium oxidizing and nirS-encoding nitrite-reducing bacteria in surface sediments of the South China Sea. Microb Ecol 66:281–296. https://doi.org/10.1007/s00248-012-0175-y
Liu K, Jiao JJ, Gu JD (2014) Investigation on bacterial community and diversity in the multilayer aquifer-aquitard system of the Pearl River Delta, China. Ecotoxicology 23(10):2041–2052
Lun YHV, Lai K-H, Wong CWY, Cheng TCE (2015) Greening and performance relativity: an application in the shipping industry. Comput Oper Res 54:295–301. https://doi.org/10.1016/j.cor.2013.06.005
Mehrdad, M, Park, H, Chandran, K, Ramalingam, K, Fillos, J, Deur, A, Beckmann, K, 2014. Biofilm population diversity and distribution in Anammox MBBR Pilot at 26 thWard WWTP in Brooklyn, New York: Molecular analysis and mathematical modelling. Proceedings of the Water Environment Federation 2014, 4605–4620. https://doi.org/10.2175/193864714815942332
Meng H, Wang Y-F, Chan H-W, Wu R-N, Gu J-D (2016) Co-occurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in subtropical acidic forest soils. Appl Microbiol Biotechnol 100:7727–7739. https://doi.org/10.1007/s00253-016-7585-6
Meng H, Yang YC, Lin JG, Denecke M, Gu JD (2017) Occurrence of anammox bacteria in a traditional full-scale wastewater treatment plant and successful inoculation for new establishment. Int Biodeterior Biodegradation 120:224–231
Mok, Y-K, 1999. A study of the future role of Hong Kong as an international port in China. https://doi.org/10.5353/th_b3195227
Mulder A, van de Graaf AA, Robertson LA, Kuenen JG (1995) Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol 16:177–183. https://doi.org/10.1111/j.1574-6941.1995.tb00281.x
Park H, Rosenthal A, Ramalingam K, Fillos J, Chandran K (2010) Linking community profiles, gene expression and N-removal in anammox bioreactors treating municipal anaerobic digestion reject water. Environ Sci Technol 44:6110–6116. https://doi.org/10.1021/es1002956
Penton CR, Devol AH, Tiedje JM (2006) Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl Environ Microbiol 72:6829–6832. https://doi.org/10.1128/AEM.01254-06
Pun, K-L, 2006. Field and model studies of water quality in Hong Kong, in: The Environment in Asia Pacific Harbours. Springer-Verlag, Berlin/Heidelberg, pp. 165–186. 10.1007/1-4020-3655-8_12
Quan Z-X, Rhee S-K, Zuo J-E, Yang Y, Bae J-W, Park JR, Lee S-T, Park Y-H (2008) Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ Microb 10:3130–3139. https://doi.org/10.1111/j.1462-2920.2008.01642.x
Ravishankara AR, Daniel JS, Portmann RW (2009) Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326:123–125. https://doi.org/10.1126/science.1176985
Rissanen, J, 1995. Shannon-Wiener information and stochastic complexity. American Mathematical Society, Providence, Rhode Island. 10.1090/psapm/052/1440920
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer KH, Wagner M (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23:93–106. https://doi.org/10.1016/S0723-2020(00)80050-8
Schmid M, Walsh K, Webb R, Rijpstra WIC, van de Pas-Schoonen K, Verbruggen MJ, Hill T, Moffett B, Fuerst J, Schouten S, Damsté JSS, Harris J, Shaw P, Jetten M, Strous M (2003) Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri,” sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 26:529–538
Schmid MC, Hooper AB, Klotz MG, Woebken D, Lam P, Kuypers MMM, Pommerening-Roeser A, Op Den Camp HJM, Jetten MSM (2008) Environmental detection of octahaem cytochrome chydroxylamine/hydrazine oxidoreductase genes of aerobic and anaerobic ammonium-oxidizing bacteria. Environ Microbiol 10:3140–3149. https://doi.org/10.1111/j.1462-2920.2008.01732.x
Schmid MC, Maas B, Dapena A, van de Pas-Schoonen K, van de Vossenberg J, Kartal B, van Niftrik L, Schmidt I, Cirpus I, Kuenen JG, Wagner M, Sinninghe Damsté JS, Kuypers M, Revsbech NP, Mendez R, Jetten MSM, Strous M (2005) Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl Environ Microbiol 71:1677–1684. https://doi.org/10.1128/AEM.71.4.1677-1684.2005
Schmid MC, Risgaard-Petersen N, van de Vossenberg J, Kuypers MMM, Lavik G, Petersen J, Hulth S, Thamdrup B, Canfield D, Dalsgaard T, Rysgaard S, Sejr MK, Strous M, Camp, den HJMO, Jetten MSM (2007) Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ Microbiol 9:1476–1484. https://doi.org/10.1111/j.1462-2920.2007.01266.x
Schubert CJ, Durisch-Kaiser E, Wehrli B, Thamdrup B, Lam P, Kuypers MMM (2006) Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ Microbiol 8:1857–1863. https://doi.org/10.1111/j.1462-2920.2006.01074.x
Seitzinger S (2008) Nitrogen cycle: out of reach. Nature 452:162–163. https://doi.org/10.1038/452162a
Shao S, Luan X, Dang H, Zhou H, Zhao Y, Liu H, Zhang Y, Dai L, Ye Y, Klotz MG (2014) Deep-sea methane seep sediments in the Okhotsk Sea sustain diverse and abundant anammox bacteria. FEMS Microbiol Ecol 87:503–516. https://doi.org/10.1111/1574-6941.12241
Shehzad A, Liu J, Yu M, Qismat S, Liu J, Zhang X-H (2016) Diversity, community composition and abundance of anammox bacteria in sediments of the North Marginal Seas of China. Microbes Environ 31:111–120. https://doi.org/10.1264/jsme2.ME15140
Shen L-D, Cheng H-X, Liu X, Li J-H, Liu Y (2017) Potential role of anammox in nitrogen removal in a freshwater reservoir, Jiulonghu Reservoir (China). Environ Sci Pollut Res Int 24:3890–3899. https://doi.org/10.1007/s11356-016-8126-7
Shen L-D, Liu S, Lou L-P, Liu W-P, Xu X-Y, Zheng P, Hu B-L (2013) Broad distribution of diverse anaerobic ammonium-oxidizing bacteria in Chinese agricultural soils. Appl Environ Microbiol 79:6167–6172. https://doi.org/10.1128/AEM.00884-13
Siegrist H, Salzgeber D, Eugster J, Joss A (2008) Anammox brings WWTP closer to energy autarky due to increased biogas production and reduced aeration energy for N-removal. Water Sci Technol 57:383–388. https://doi.org/10.2166/wst.2008.048
Stevens, H, 1999. The Port of Hong Kong, in: The Institutional Position of Seaports, GeoJournal Library. Springer Netherlands, Dordrecht, pp. 229–253. https://doi.org/10.1007/978-0-585-35323-4_9
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Médigue C, Collingro A, Snel B, Dutilh BE, Op Den Camp HJM, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes H-W, Weissenbach J, Jetten MSM, Wagner M, Le Paslier D (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790–794. https://doi.org/10.1038/nature04647
Sun W, Xia C, Xu M, Guo J, Wang A, Sun G (2014a) Diversity and distribution of planktonic anaerobic ammonium-oxidizing bacteria in the Dongjiang River, China. Microbiol Res 169:897–906. https://doi.org/10.1016/j.micres.2014.05.003
Sun W, Xu M-Y, Wu W-M, Guo J, Xia C-Y, Sun G-P, Wang A-J (2014b) Molecular diversity and distribution of anammox community in sediments of the Dongjiang River, a drinking water source of Hong Kong. J Appl Microbiol 116:464–476. https://doi.org/10.1111/jam.12367
Thamdrup B, Dalsgaard T (2002) Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl Environ Microbiol 68:1312–1318. https://doi.org/10.1128/AEM.68.3.1312-1318.2002
van de Graaf AA, Mulder A, de Bruijn P, Jetten MS, Robertson LA, Kuenen JG (1995) Anaerobic oxidation of ammonium is a biologically mediated process. Appl Environ Microbiol 61:1246–1251
Van Duc L, Song B, Ito H, Hama T, Otani M, Kawagoshi Y (2017) High growth potential and nitrogen removal performance of marine anammox bacteria in shrimp-aquaculture sediment. Chemosphere 196:69–77. https://doi.org/10.1016/j.chemosphere.2017.12.159
Wan Z, Zhu M, Chen S, Sperling D (2016) Pollution: three steps to a green shipping industry. Nature 530:275–277. https://doi.org/10.1038/530275a
Wang J, Gu J-D (2013) Dominance of Candidatus Scalindua species in anammox community revealed in soils with different duration of rice paddy cultivation in Northeast China. Appl Microbiol Biotechnol 97:1785–1798. https://doi.org/10.1007/s00253-012-4036-x
Wang Y-F, Gu J-D (2012) Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl Microbiol Biotechnol 97:7015–7033. https://doi.org/10.1007/s00253-012-4430-4
Wang YF, Gu JD (2014) Effects of allylthiourea, salinity, and pH on ammonia/ammonium-oxidizing prokaryotes in mangrove sediment incubated in laboratory microcosms. Appl Microbiol Biotechnol 98(7):3257–3274
Wang S, Zhu G, Peng Y, Jetten MSM, Yin C (2012a) Anammox Bacterial Abundance, Activity, and Contribution in Riparian Sediments of the Pearl River Estuary. Environ Sci Technol 46:8834–8842. https://doi.org/10.1021/es3017446
Wang Y, Zhu G, Harhangi HR, Zhu B, Jetten MSM, Yin C, Op Den Camp HJM (2012b) Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS Microbiol Lett 336:79–88. https://doi.org/10.1111/j.1574-6968.2012.02654.x
Wang YF, Li XY, Gu JD (2014) Differential responses of ammonia/ammonium-oxidizing microorganisms in mangrove sediment to amendment of acetate and leaf litter. Appl Microbiol Biotechnol 98(7):3165–3180
Wang Y-F, Feng Y-Y, Ma X, Gu J-D (2013) Seasonal dynamics of ammonia/ammonium-oxidizing prokaryotes in oxic and anoxic wetland sediments of subtropical coastal mangrove. Appl Microbiol Biotechnol 97:7919–7934. https://doi.org/10.1007/s00253-012-4510-5
Yang X-R, Li H, Nie S-A, Su J-Q, Weng B-S, Zhu G-B, Yao H-Y, Gilbert JA, Zhu Y-G (2015) Potential contribution of anammox to nitrogen loss from paddy soils in southern China. Appl Environ Microbiol 81:938–947. https://doi.org/10.1128/AEM.02664-14
Yang Y, Li M, Li X-Y, Gu J-D (2018) Two identical copies of the hydrazine synthase gene clusters found in the genomes of anammox bacteria. Int Biodeterior Biodegradation 132:236–240. https://doi.org/10.1016/j.ibiod.2018.04.011
Yoshinaga I, Amano T, Yamagishi T, Okada K, Ueda S, Sako Y, Suwa Y (2011) Distribution and Diversity of Anaerobic Ammonium Oxidation (Anammox) Bacteria in the Sediment of a Eutrophic Freshwater Lake, Lake Kitaura, Japan. Microbes Environ 26:189–197. https://doi.org/10.1264/jsme2.ME10184
Zeng X-L, Liu X-G, Wu Z-F, Shi X, Lu S-M (2016) [Community Characteristics of ANAMMOX Bacteria in Subsurface Flow Constructed Wetland (SSFCW) for Processing of Aquaculture Waster Water]. Huan Jing Ke Xue 37:615–621
Zhang Y, Ruan X-H, Op Den Camp HJM, Smits TJM, Jetten MSM, Schmid MC (2007) Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China). Environ Microbiol 9:2375–2382. https://doi.org/10.1111/j.1462-2920.2007.01357.x
Zhao S, Zhuang L, Wang C, Li Y, Wang S, Zhu G (2017) High-throughput analysis of anammox bacteria in wetland and dryland soils along the altitudinal gradient in Qinghai-Tibet Plateau. Microbiologyopen 41:e00556. https://doi.org/10.1002/mbo3.556
Zhijie F (1988) Pollution causes concern in South China Sea. Mar Pollut Bull 19:92. https://doi.org/10.1016/0025-326X(88)90696-0
Zhou Z, Chen J, Meng H, Dvornyk V, Gu J-D (2017) New PCR primers targeting hydrazine synthase and cytochrome c biogenesis proteins in anammox bacteria. Appl Microbiol Biotechnol 101:1267–1287. https://doi.org/10.1007/s00253-016-8013-7
Zhou Z, Wei Q, Yang Y, Li M, Gu J-D (2018) Practical applications of PCR primers in detection of anammox bacteria effectively from different types of samples. Appl Microbiol Biotechnol 102(14):5859–5871
Zhu G, Wang S, Feng X, Fan G, Jetten MSM, Yin C (2011) Anammox bacterial abundance, biodiversity and activity in a constructed wetland. Environ Sci Technol 45:9951–9958. https://doi.org/10.1021/es202183w
Acknowledgements
This project was supported by Hong Kong PhD Fellowship (XWZ), Natural Science Foundation of China (Grant No. 92051103, 41907202) (J-DG, HM), Shenzhen Scientific Project Fund (No. JCYJ20170410103015603)(WSL), RGC TRS Project T21-711/16-R (XYL).
Funding
This study was funded by Natural Science Foundation of China (Grant No. 92051103, 41907202) (J-DG, HM), The Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No. 19KJB180004) (HM), Shenzhen Scientific Project Fund (No. JCYJ20170410103015603) (WSL), and TRS Project T21-711/16-R and a Hong Kong PhD Fellowship (XWZ).
Author Contribution
XZ and J-DG conceived and designed the research. XZ, HM, and YY conducted experiments. PL coordinated the sampling and logistics on transportation. YL and CU contributed to the interpretation of results and discussion. WL, PL, XY and J-DG provided the fund for this research. XZ, HM, YY, WW, and J-DG analyzed data and wrote the first draft of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
All authors participate, read and approved this manuscript.
Animal research
This research does not contain any studies with human participants or animals performed by any of the authors.
Plant reproducibility
This study did not involve any plants or plant materials.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, X., Meng, H., Yang, Y. et al. Diversity, abundance, and distribution of anammox bacteria in shipping channel sediment of Hong Kong by analysis of DNA and RNA. Ecotoxicology 30, 1705–1718 (2021). https://doi.org/10.1007/s10646-020-02332-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-020-02332-y